Figure 1.
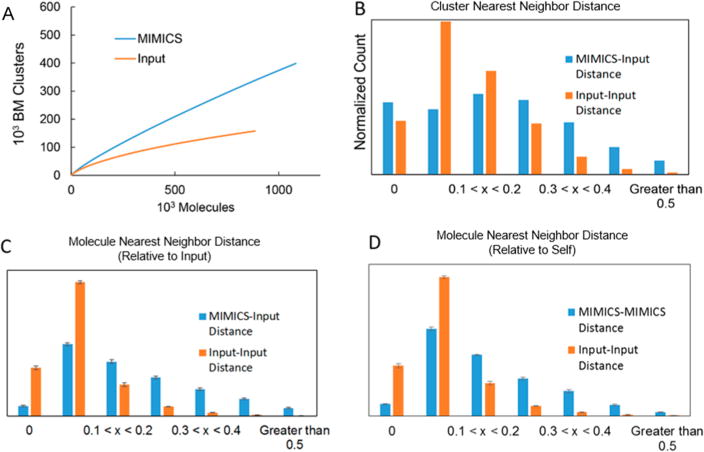
Structural novelty comparison. (A) Bemis–Murcko clustering8 was conducted on the MIMICS and input molecule sets to assess the diversity and novelty of central structural motifs. The number of unique scaffolds produced as a function of MIMICS molecules generated is displayed. (B) The Tanimoto distance between a particular structure and its nearest neighbor in the input set was computed using the Open Babel7 FP2 fingerprint for samples of MIMICS and input molecules. (C) Nearest-neighbor distance histogram for MIMICS molecules and input molecules relative to the input. (D) Nearest-neighbor distance histogram for MIMICS molecules and input molecules relative to themselves.
