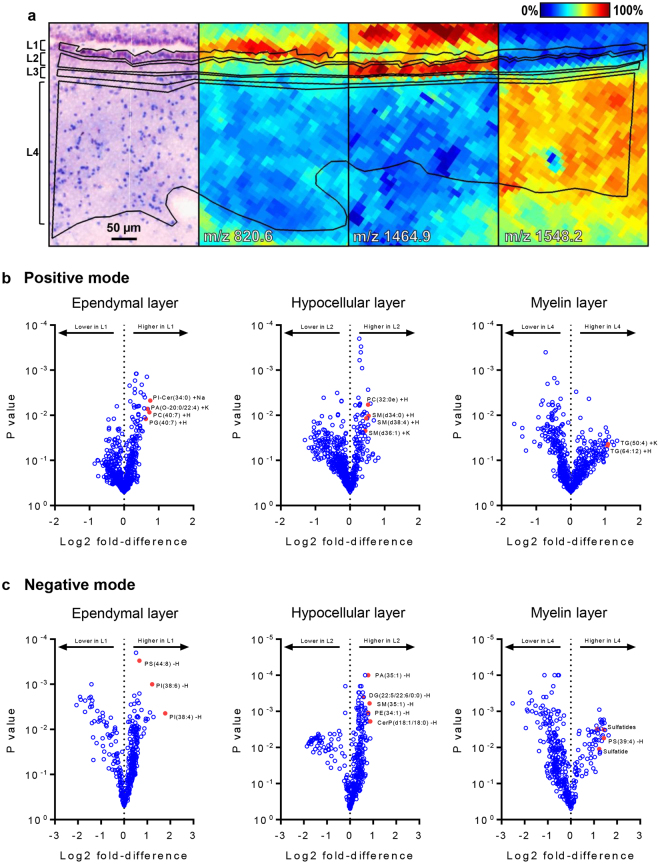
Figure 4.
The lipidomic architecture of the constituent layers of the SVZ. (a) Histology-guided delineation of the four SVZ layers as analysis regions in SCiLS Lab software, with example m/z signals shown that were found to co-localise with the ependymal layer, hypocellular layer or the myelin layer. Scale bar: 50 µm. (b,c) Volcano plots of individual m/z signals in positive and negative polarity with log2 transformed fold differences plotted against P-values arising from one-sample t-tests. Summary spectra for each layer (including the astrocytic ribbon) were obtained and log2 fold-differences and t-test P-values derived by comparing, for each m/z signal, its intensity value in the summary spectrum for each layer with the mean intensity of the summary spectra corresponding to the three other SVZ layers. L1 = ependymal layer; L3 = hypocellular layer; L4 = myelin layer. Lipid species that are discussed in the text are arbitrarily marked in red here and the assignments for these are indicated.