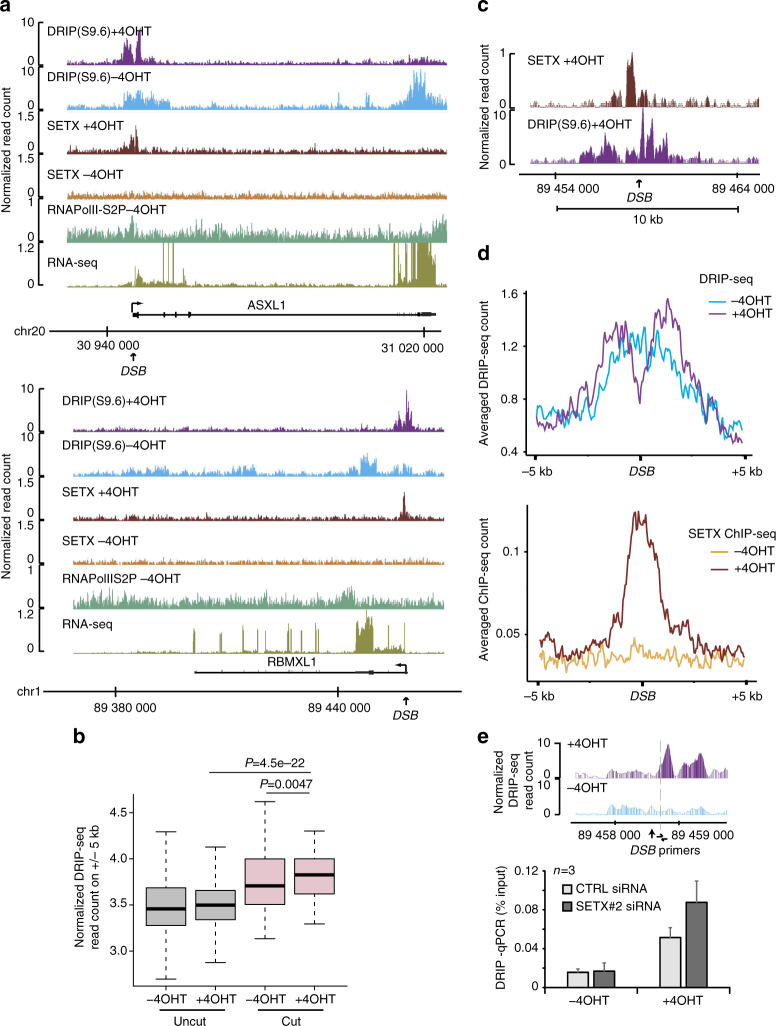
Fig. 2.
Senataxin removes DSB-induced RNA:DNA hybrids in active loci. a Genome browser screenshots representing DRIP-seq reads count before (−4OHT) and after damage induction (+4OHT) at two individual AsiSI sites. Senataxin profiles in both conditions are also shown, together with BLESS enrichment following DSB induction as well as RNA PolII-S2P and RNA-seq prior to DSB induction. b Box plots representing DRIP-seq reads count before (−4OHT) and after (+4OHT) DSB induction at sites displaying AsiSI-induced cleavage (“cut”) or not (“uncut”). Center line: median; box limits: 1st and 3rd quartiles; whiskers: maximum and minimum without outliers. P values are indicated (Wilcoxon–Mann–Whitney test). c Close-up genome browser screenshot of senataxin and RNA:DNA hybrids at an individual DSB upon DSB induction. d Average DRIP-seq (top) and senataxin ChIP-seq (bottom) profiles on a ±5 kb window centered on the 80 AsiSI induced DSBs. e DRIP-qPCR in control (CTRL) and senataxin (SETX#2) depleted cells before (−4OHT) and after (+4OHT) DSB induction in DIvA cells as indicated. The position of the primers used to quantify RNA:DNA hybrids by qPCR are indicated on the genome browser screenshot above. Mean and s.e.m. of three biological replicates is shown