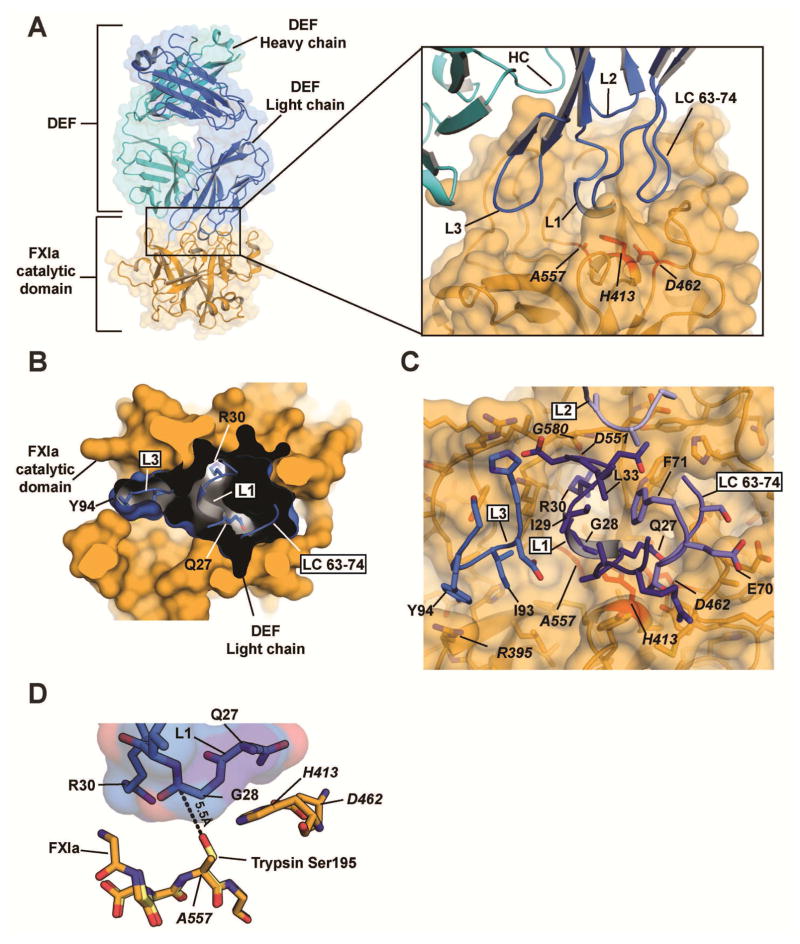
Figure 2. Structure of the DEF-FXIa catalytic domain complex.
(A) Cartoon and surface representation of the DEF-FXIa catalytic domain complex. DEF Heavy chain (cyan) and Light chain (marine) are indicated. Inset shows CDR interactions with FXIa (light orange). Catalytic triad residues are red. (B) Cutaway view showing DEF light chain engagement with the active site. Black shows the DEF light chain interior. DEF light chain loops are shown and labeled. Select DEF residues are indicated.
(C) Details of the DEF-FXIa interaction. FXIa residues are indicated by italics.
(D) Orientation of DEF CDR L1 loop residues relative to the FXIa catalytic triad. The structure of trypsin (PDB code 1PQA, light yellow) was superimposed on that of FXIaiPD (light orange) to predict the location of the oxygen nucleophile in the active site serine of FXIa (an alanine in FXIaiPD, introduced to prevent autolysis during protein concentration). The distance between this location and Gly28 in DEF CDR L1 is shown.