Abstract
Nonsense-mediated mRNA decay, or NMD, is a quality control mechanism that identifies cytoplasmic mRNAs containing translational termination (stop) codons in specific contexts—either premature termination codons or unusually long 3΄ untranslated regions (UTRs)—and targets them for degradation. In recent studies, researchers in different labs have knocked out important genes involved in NMD, the up-frameshift genes Upf2 and Upf3a, and one component of chromatoid bodies, the Tudor domain-containing protein Tdrd6, and examined the consequences for spermatogenesis. Disruption of Upf2 during early stages of spermatogenesis resulted in disappearance of nearly all spermatogenic cells through loss of NMD. However, disruption of Upf2 during postmeiotic stages resulted in decreased long 3΄ UTR-mediated NMD but no interruption of exon junction-associated NMD. This difference in NMD targeting is possibly due to increased expression of Upf3a in postmeiotic germ cells that antagonizes the functions of Upf3b and somehow favors long 3΄ UTR-mediated NMD. Tying these all together, loss of Tdrd6, a structural component of the germ cell-specific cytoplasmic structures called chromatoid bodies, also resulted in loss of long 3΄ UTR-mediated NMD by interfering with UPF1/UPF2 interactions, delocalizing UPF1, and destroying chromatoid body integrity. These results suggest that chromatoid bodies play a specialized role in modulating the NMD machinery in postmeiotic spermatids.
Keywords: nonsense-mediated decay, premature stop codon, spermatogenesis, testis, Sertoli cell, mRNA processing, 3΄ untranslated region, chromatoid body
Summary Sentence
Nonsense-mediated decay is how cells eliminate mRNAs having premature stop codons or long 3΄ untranslated regions. Recent findings in male germ cells uncover new mechanisms for nonsense-mediated decay that shine light on existing models.
Abbreviations
- Amh-Cre
mice expressing the Cre recombinase under the control of the anti-Müllerian hormone promoter in Sertoli cells starting before birth (∼E12.5)
- CB
chromatoid body, a nonmembranous perinuclear cytoplasmic structure observed in round and elongating spermatids, composed of multiple RNA species (including mRNAs, miRNAs, piRNAs), RNA transport proteins (such as KIF7b), and miRNA and piRNA biogenesis proteins (Argonaute proteins, Dicer, Tudor domain proteins, etc.); the work reviewed here also suggests a role for CBs in NMD
- Ddx4-Cre
mice expressing the Cre recombinase under the control of the DEAD-Box Helicase 4/VASA homolog promoter in primordial germ cells before birth (∼E15.5)
- E9.5, E12.5, etc.
embryonic day 9.5, 12.5, etc.
- eIF4E
eukaryotic translational initiation factor 4E cap-binding protein
- EJC
the nuclear exon junction complex, comprised of eIF4A3, Magoh, RBM8A, and CASC3
- eRF1, eRF3
eukaryotic translational release factors 1, 3
- IMC
intermitochondrial cement, a nonmembranous perinuclear cytoplasmic structure observed in spermatocytes, usually associated with mitochondrial clusters; IMCs become CBs in spermatids after meiosis
- MSCI
meiotic sex chromosome inactivation
- MVH
mouse VASA homolog, encoded by the Ddx4 gene
- NMD
nonsense-mediated decay
- ORF
open reading frame
- P5, P60, etc.
Postnatal day 5, 60, etc.
- PABPC1
poly(A) binding protein, cytoplasmic
- PABPN1
poly(A) binding protein, nuclear
- PTC
premature termination (stop) codon
- Sec
selenocysteine
- SMG1, SMG6, SMG7, etc.
suppressor with morphogenic effect on genitalia 1, 6, 7, etc.
- Stra8-Cre
mice expressing the Cre recombinase under the control of the stimulated by Retinoic acid 8 promoter in meiotic and haploid male germ cells (starting at ∼P3)
- SURF complex
SMG1C/UPF1/eRF1/eRF3 cytoplasmic complex
- TDRD6
Tudor domain-containing protein 6
- Upf1, Upf2, Upf3a, Upf3b
mouse homologs of up-frameshift genes that were first identified in Saccharomyces cerevisiae; the proteins are referred to as UPF1, UPF2, etc.
- UTR
mRNA untranslated region, e.g., the 3΄ UTR
Introduction
Nothing is so simple that its tenets cannot be challenged in testis biology. New results from several labs focused on knockouts of key NMD protein factors in testis in mice and thus challenged the mechanisms of nonsense-mediated mRNA decay (NMD) in male germ cells. Nonsense-mediated mRNA decay is the cytoplasmic process by which mRNAs containing “errors”—improper splicing, transcriptional errors, mutations that result in premature stop codons, and more—are eliminated from the cytoplasmic pool of translated mRNAs. Nonsense-mediated mRNA decay also eliminates mRNAs that arise through nonerroneous mechanisms that result in translational stop codons in certain premature contexts: alternative splicing, alternative polyadenylation, small upstream open reading frames (ORFs), or low cellular selenium (Figure 1). In a felix culpa from testis biology, the rules governing NMD are illuminated in male germ cells and their associated somatic cell populations, yielding a better understanding of both male reproduction and mRNA quality control. In this review, we evaluate recent mouse knockout models of two components of NMD (Upf2 [1,2] and Upf3a [3]) and one component of the chromatoid body (CB; Tdrd6 [4]) in male germ and Sertoli cells. Together, these manuscripts argue for two distinct modes for NMD: one mode (in somatic cells such as Sertoli cells) in which NMD is mostly active on mRNAs containing premature termination codons in alternatively spliced exons, and a second mode (in male germ cells) in which NMD is managed by CBs, thus acting on mRNAs containing long 3΄ untranslated regions (UTRs). Separation of these modes in male germ cells helps to better understand NMD mechanisms in all cells.
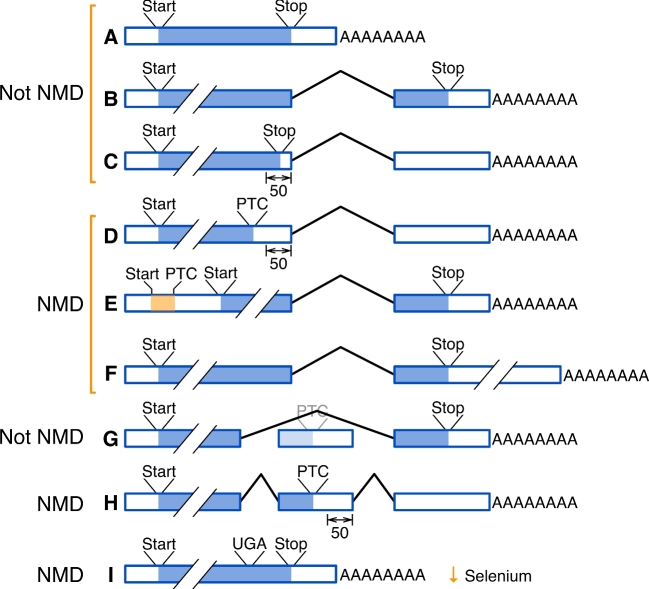
Figure 1.
Many different mRNA errors that cause premature termination/stop codons (PTCs) can lead to nonsense-mediated decay (NMD). Boxes represent exons, lines represent introns, ORFs are in blue, and Start (AUG) or Stop (UAA, UAG, or UGA) codons are as indicated. (A–C) Most mRNAs are not subject to NMD. (A) Messenger RNAs from genes lacking introns, or (B) with ORFs resulting in stop codons in the last exon (i.e., downstream of the last intron) are not subject to NMD. (C) The major exception to the last exon rule is mRNAs with the stop codon fewer than 50 nucleotides upstream of the last intron. (D–F) Nonsense-mediated decay is invoked for mRNAs with the stop codon (D) greater than 50 nucleotides upstream of the last exon, (E) with a short ORF in the 5΄ UTR, or (F) in the last exon, but followed by a long (>350 nt) 3΄ untranslated region (UTR). Nonsense-mediated mRNA decay in these groups might be invoked by genomic mutations, by cotranscriptional errors, or by post-transcriptional mechanisms (say, alternative polyadenylation). (G, H) Alternative splicing can insert a stop codon to invoke NMD. (G) In this example, exon skipping omits an exon with an NMD-inducing stop codon, but exon inclusion will include that exon with the offending stop codon, resulting in NMD. (I) In some mRNAs, selenocysteine is encoded by UGA, which is also a stop codon. In conditions of low selenium, the Sec-tRNA is in low abundance, triggering NMD for selenoprotein mRNAs.
Nonsense-mediated decay degrades mRNAs with premature stop codons or anomalously long 3΄ untranslated regions
Nonsense-mediated decay is the mechanism that targets mRNAs for rapid degradation if they contain premature termination or “stop” codons (PTCs). A premature termination codon is defined as an in-frame translational stop codon that is more than 50 nucleotides upstream of the last (3΄-most) exon in the gene, although there are other contexts as well (Figure 1). First discovered in yeast [5,6], NMD was quickly determined to be a universal feature of eukaryotic mRNA biosynthesis that is invoked when mRNAs fail to pass “quality control” standards with regard to their translational ORFs. Of the several mechanisms by which a PTC can be introduced into an mRNA, the most common is alternative splicing [7], which is also a frequent mechanism of gene regulation in male germ cells [8–10].
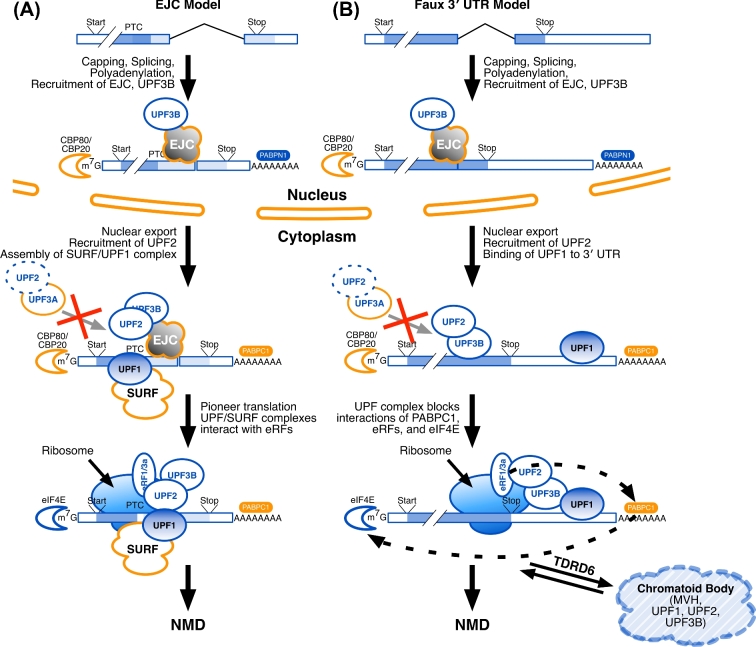
If NMD detects mRNA stop codons (which are defined by translation in the cytoplasm), then how can it tell when the stop codon is in the last exon (which is defined by splicing in the nucleus)? For many mRNAs, the answer is that they are “marked” while still in the nucleus with proteins that bind near exon junctions, including the junction of the penultimate and last exons [11,12]. These protein marks are carried with the nascent mRNA into the cytoplasm during nuclear export, and consist of the exon junction complex (EJC), UPF3B, and other components (Figure 2). Once in the cytoplasm, the protein marks recruit additional proteins (including UPF1 and UPF2) that govern interactions with ribosomes, translational termination factors, and the cytoplasmic poly(A)-binding protein PABPC1. Consequently, mRNAs with PTCs are recognized during early “pioneer” rounds of translation to be eliminated from the translational pool and degraded (Figure 2A). In mammals, the degradation involves the proteins SMG5, SMG6, and SMG7 [13].
Figure 2.
Several models vie to account for nonsense-mediated decay (NMD) in different cell types. (A) In the Exon Junction Complex (EJC) model, after splicing, EJC proteins bind at the junctions of the last and penultimate exons, to be joined by UPF3B (the EJC “marks”). This nucleoprotein complex is then exported from the nucleus to the cytoplasm, where it is joined by UPF2, the SMG1C/UPF1/eRF1/eRF3 (SURF) complex proteins, and UPF1. Nuclear poly(A) binding protein (PABPN1) is replaced by its cytoplasmic equivalent (PABPC1), and the nuclear cap-binding proteins (CBP80, CBP20) are replaced by their translational equivalents, including eIF4E. A pioneer round of translation ensues, and translational release factors (eRF1 and eRF3a) allow the ribosome to recognize the stop codon. If the stop codon is premature, that mRNA is targeted for degradation. (B) The Faux 3΄ UTR model is similar to the EJC model in its nuclear marking steps. In the cytoplasm, UPF2 binds to UPF3B. UPF1 binds to multiple RNA sites within the 3΄ UTR. In a way that is not yet understood, multiple copies of UPF1 (as might bind to a long 3΄ UTR) will interfere with an interaction between the release factors (eRF1, eRF3a) and PABPC1; multiple copies of UPF1 might also interfere with the interaction of PABPC1 and the cytoplasmic cap-binding proteins (eIF4E and others). In male germ cells, TDRD6 acts to localize UPF1, UPF2, and other factors to the CBs in postmeiotic spermatids. Also in both models (A) and (B), UPF3A may bind to UPF2 in the cytoplasm to obstruct the UPF2-UPF3B interaction and suppress EJC-stimulated NMD.
It is less well understood how mRNAs with long (>350 nt) 3΄ UTRs (Figure 1F) are targeted for NMD. This mechanism seems to involve the distance between the termination codon and the poly(A)-binding protein (Figure 2B), and requires UPF1, SMG6, and SMG7, and other proteins [12,14]. The likely mechanism involves binding of multiple copies of UPF1 to the mRNA throughout the 3΄ UTR (Figure 2B). In somatic cells, it is known that specific mRNAs can evade this targeting if they contain A/U-rich sequences in their 3΄ UTRs [15]. Because of alternative cleavage and polyadenylation, 3΄ UTRs of any given gene can differ in length[16–18]. Alternative cleavage and polyadenylation is particularly important in controlling male germ cell gene expression [10,19,20]. As a consequence, 3΄ UTRs of testis-specific genes become progressively shorter during spermatogenesis [20–22], a situation that is reminiscent of the 3΄ UTR shortening seen in cancer cells [23].
In further precedent for 3΄ UTRs playing roles in controlling germ cell mRNA expression, the polypyrimidine tract binding protein 1 is involved in protecting mRNAs with long 3΄ UTRs by interfering with binding of UPF1 near the stop codon [24]. Its paralog, Ptbp2, is essential for male germ cell development through its effects on splicing [9] and has been implicated in stabilizing mRNAs with long 3΄ UTRs [25]. The model emerging is that mRNAs with long 3΄ UTRs are often targeted for degradation in postmeiotic germ cells, but that specific mRNAs can be marked to evade degradation. These points suggest that mechanisms of NMD play important roles in male fertility, as confirmed in the manuscripts reviewed here.
Upf2 is required for mouse Sertoli cell development and male fertility, and eliminates mRNAs with premature termination codons
Weischenfeldt and colleagues in Porse's laboratory examined conditional Cre-Lox truncation of Upf2 that produced a shorter, functionally inactive form of the UPF2 protein in mice [26,27]. Global truncation of Upf2 in all tissues was lethal before embryonic day 9.5 (E9.5). Further, truncation of Upf2 specifically in immature T cells resulted in the death of hematopoietic stem cells. In these cells, the loss of UPF2, a key component of the NMD pathway [7,28], resulted in an increased abundance of alternatively spliced transcripts containing PTCs. Interestingly, Upf2 ablation in the granulocyte/monocyte lineage was tolerated, suggesting that NMD was not critical in those more differentiated cell types.
To extend this work to male reproduction, Porse's and Yan's laboratories joined efforts to examine Upf2 ablation in Sertoli [1] and germ cells [2] using the conditional knockout animals generated earlier in Porse's laboratory. In the first study, Bao et al. [1] chose to examine effects of Upf2 truncation in Sertoli cells, somatic cells that provide supporting functions to germ cells during spermatogenesis. By crossing Upf2fl/fl mice with mice carrying the Cre recombinase under the control of the anti-Müllerian hormone promoter (Amh-Cre), these authors inactivated Upf2 in Sertoli cells beginning before birth. These mice were infertile and had significantly smaller testes than mice with intact Upf2. By postpartum day 60 (P60), the seminiferous tubules were largely devoid of germ cells with only Leydig cells present (a Leydig cell-only phenotype). The authors concluded that truncation of Upf2 led to failure of the Sertoli cells to differentiate and thus were unable to nurture normal germ cell development during the first wave of spermatogenesis.
Disrupted mRNA processing in Upf2-truncated Sertoli cells
At P4 (a time chosen because it preceded the changes in other testicular cell types), high-throughput RNA sequencing demonstrated that similar numbers of transcripts were either increased (∼1200) or decreased (∼1400) in the Upf2-truncated Sertoli cells compared to Upf2-intact cells [1]. Many of these affected mRNAs were linked to RNA splicing and processing. About 30% of the upregulated genes in Upf2-truncated Sertoli cells contained premature termination codons, consistent with the established role of Upf2 in NMD in somatic cells.
Alternative splicing is also a prominent regulator of male meiosis [8,29]. Nine core splicing factors were deregulated in Upf2-truncated Sertoli cells [1], decreasing the major isoforms of two alternatively spliced transcription factors, Wt1 and Dmrt1 that are responsible for Sertoli cell differentiation. Many other gene products necessary for Sertoli cell differentiation were also mis-spliced or otherwise downregulated. Ultimately, these errors resulted in failure of Sertoli cell differentiation and the infertile phenotype.
In germ cells, Upf2-mediated nonsense-mediated decay selectively eliminates mRNAs with long 3΄ untranslated regions
In a second study using the same Upf2fl/fl allele as above, Bao et al. examined the role of Upf2 in male germ cells [2]. Crossing Upf2fl/fl mice with mice-expressing Cre recombinase prenatally in primordial germ cells (Ddx4-Cre mice [30]) resulted in seminiferous tubules containing mostly Sertoli cells and few, if any, germ cells, confirming that NMD is necessary for early prospermatogonial germ cell development.
These results prompted the authors to look at roles for Upf2 during postnatal stages of spermatogenesis by breeding Upf2fl/fl mice with Stra8-Cre mice, resulting in Upf2 truncation in early-stage spermatogonia and spermatocytes [31]. The resulting Upf2 conditional knockout mice had smaller testes due to defects in spermatogenesis, including delayed meiosis, reductions in spermatocytes and spermatids, and large vacuoles with multinucleated giant cells in the seminiferous tubules [2]. Clearly, loss of NMD function was also deleterious for male gamete development when Upf2 was truncated in these later stages of spermatogenesis.
The surprising finding, though, was how the loss of Upf2 caused those problems. The authors determined that very few (∼7%) of the upregulated alternatively spliced mRNAs contained PTCs in Upf2-truncated testes [2]; in other words, loss of Upf2 function did not affect traditional EJC-associated (herein “EJC-stimulated”) NMD (Figure 1G and H) in postmeiotic male germ cells. Instead, there was an accumulation of mRNAs with long 3΄ UTRs (Figure 1F) among the population of genes with multiple transcripts (i.e., mRNAs that were alternatively spliced, polyadenylated, or both). In germ cells, the mechanisms of EJC-stimulated NMD were less evident while the mechanisms of long 3΄ UTR-mediated NMD remained prominent. This suggests the formal possibility that UPF2 is not required for NMD in male germ cells. However, the more likely explanation is that UPF2 is functionally sequestered in germ cells. As discussed below, this sequestration might involve degradation of mRNAs with long 3΄ UTRs which is associated with RNA-processing centers called CBs that are unique to postmeiotic spermatids (Figure 3).
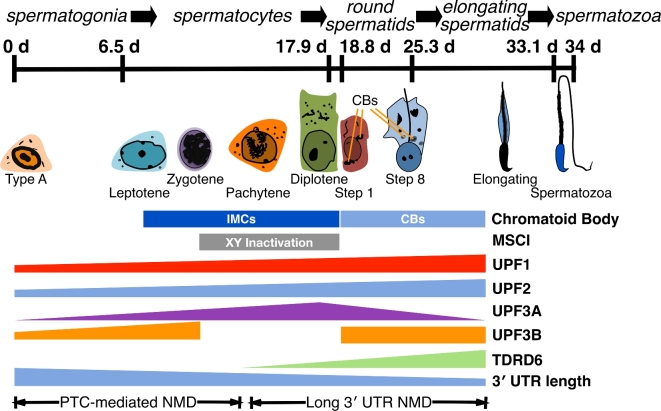
Figure 3.
Expression of NMD components during spermatogenesis. Spermatogenesis takes about 34 days in mice (top). Cell types progress from spermatogonia (that still undergo mitosis) through multiple stages of meiosis (spermatocytes), through postmeiotic differentiation stages (step 1 through step 8 round spermatids and elongating spermatids), and emerge as spermatozoa. Spermatozoa must traverse the epididymis to become competent for fertilization (not shown). In spermatocytes, cloud-like (“nuage”) structures called the intermitochondrial cement (IMC) assemble near mitochondria. These structures coalesce into perinuclear structures called chromatoid bodies (CBs) after meiosis. In the absence of an enclosing membrane, proteins such as the Tudor domain-containing TDRD6 act as organizational components of the CB; UPF1, UPF2, and UPF3B are also found there. Also during meiosis, the X and Y chromosomes are inactivated transcriptionally and sequestered into sex bodies, a process known as meiotic sex chromosome inactivation (MCSI). Thus, X-linked genes (such as Upf3b) are inactivated during meiosis while autosomal paralogs (Upf3a) are expressed. Several studies have shown that 3΄ UTR length of mRNAs becomes shorter as spermatogenesis decreases. The studies reviewed here indicate that long 3΄ UTR-mediated NMD is prominent during postmeiotic stages. We speculate that EJC-stimulated NMD is prominent during earlier stages of spermatogenesis, although the studies did not address this point directly.
Two homologs, Upf3a and Upf3b, compete antagonistically for control of nonsense-mediated decay in male germ cells
How might one mechanism of NMD be reduced in male germ cells (EJC-stimulated NMD) while the other mechanism (long 3΄ UTRs) remains active? A study from Wilkinson's lab, Shum et al. [3] suggest a solution involving functions of Upf3a and Upf3b. In humans, the UPF3A and UPF3B mRNAs are expressed in all tissues, although the UPF3A mRNA is markedly more abundant in testis [32]. Interestingly, UPF3B regulates levels of the UPF3A mRNA through EJC-stimulated NMD such that the UPF3B protein is predominantly expressed in most adult tissues [33]. The UPF3B gene (located on the X chromosome) and the UPF3A gene (located on an autosome) are paralogs: they arose by gene duplication in an ancestor of modern vertebrates [3,34]. In eutherian mammals, such gene duplicates are often pressed into service, especially if one of them is on the X chromosome. During mammalian meiosis, the X and Y chromosomes are silenced, an epigenetic phenomenon known as meiotic sex chromosome inactivation (MSCI [35–37]). Consequently, if an essential gene on the X chromosome is inactivated (e.g., UPF3B), an autosomal paralog is deployed during pachynema (e.g., UPF3A). Indeed, mutations in UPF3B can cause an X-linked intellectual disability in humans [38].
UPF3B binds to both the EJC and UPF2 in the cytoplasm [39,40], bridging these proteins to enable NMD (Figure 2). Shum et al. [3] demonstrate that, while the UPF3B protein enables NMD, UPF3A is an NMD repressor, probably acting to mask UPF2 and prevent its interaction with the EJC. During spermatogenesis, the UPF3B protein is expressed in premeiotic germ cells, but its expression is shut down in meiotic cells due to MSCI (Figure 3). At that point, UPF3A expression increases so that it is prominent in spermatocytes. In postmeiotic germ cells, UPF3A expression is still evident while UPF3B expression returns, so both forms are expressed in spermatids. Interestingly, UPF3A expression is diffuse in the cytoplasm of round spermatids, while UPF3B expression is localized to cytoplasmic granules reminiscent of CBs ([3], and M. Wilkinson and L. Huang, personal communication). This suggests different availabilities of UPF3A and UPF3B for NMD in these cells, and possibly implicates CBs in NMD (see below).
Shum et al. [3] created conditional deletions of Upf3a in mouse cells and tissues. In neural stem cells, knockdown of UPF3A appeared to destabilize a subset of mRNAs with longer 3΄ UTRs while stabilizing a different subset, consistent with an antagonistic role for UPF3A in NMD. Perhaps unsurprisingly, Upf3a ablation in early embryos was lethal between E4.5 and E8.5 demonstrating that its antagonistic functions are nonetheless important for gene expression in early development.
The authors next examined Upf3a ablation in mouse germ cells. Upf3afl/fl mice mated with Stra8-Cre mice resulted in male offspring with Upf3a ablated in spermatogonia. This resulted in reduced sperm counts and seminiferous tubules showing loss of later stage germ cells, reminiscent of that in the Upf2-truncated mice from Bao et al. [2]. Analysis of several NMD-substrate mRNAs in the Upf3a-ablated mouse testes demonstrated that PTC-containing messages arising from alternatively spliced mRNAs were reduced [3], consistent with UPF3A acting as a suppressor of that branch of NMD. Shum et al. [3] do not directly assess effects of Upf3a on mRNAs with long 3΄ UTRs in spermatids, but their findings in P19 cells and spermatocytes support UPF3A stabilization of mRNAs with long 3΄ UTRs.
Together, we speculate that these results suggest an explanation of how Upf2 inactivation or disruption in postmeiotic germ cells acts largely on mRNAs with long 3΄ UTRs but not on traditional EJC-stimulated transcripts [2]. Generally, in nongerm cells (such as Sertoli cells [1]), UPF3B is more abundant than UPF3A, enabling the elimination of PTC-containing mRNAs through interaction between UPF2 and UPF3B (Figure 2A). In meiotic and postmeiotic germ cells, on the other hand, UPF3A is more abundant [3], and thus could suppress the EJC-stimulated effects by blocking UPF2 (Figure 2B) and favoring long 3΄ UTR surveillance. Incidental support for this proposal comes from the observation that the piRNA machinery (which is abundant in CBs) can act to cleave mRNAs before degradation [41]. This illustrates the multiple mRNA catabolic processes that are active in CBs, some of which might use overlapping machinery.
Chromatoid bodies are depots for nonsense-mediated decay components in postmeiotic germ cells, and their disruption disables decay of mRNAs with long 3΄ untranslated regions
The true functions of CBs remain mysterious. Chromatoid bodies are perinuclear cytoplasmic granules in postmeiotic spermatids that arise from the intermitochondrial cement structures seen during meiosis (Figure 3). As such, they resemble P-bodies or stress granules in other cell types [42]. Chromatoid bodies seem to be depots for many types of RNAs: mRNAs, miRNAs, piRNAs, and others [43]. They are also depots for many cytoplasmic RNA-binding proteins [42] including those involved in miRNA processing (AGO1, AGO2, Dicer), translation (PABPC1 and PABPC2 [44]), piRNA processing (MILI [45]), retroposon-suppressing components (TDRD5 [46]), and other Tudor domain-containing proteins (TDRD1 [45] and TDRD6 [47]). Somatic cells have many of these RNA metabolic functions, but they do not require these unique cytoplasmic structures, which is part of the mystery of CBs in germ cells.
Both Bao et al. [2] and Fanourgakis et al. [4] noted that, in round spermatids, the majority of UPF2 was clustered in CBs. Both groups of authors suggest that the CB is a germ cell-specific site of RNA catabolism, wherein UPF2 plays a central role in NMD. Although Shum et al. [3] did not address it directly in their manuscript, examination of their UBF3B stained images reveals that the protein is localized in speckles resembling CBs in spermatids (M. Wilkinson and L. Huang, personal communication). In contrast, UPF3A was generally cytoplasmic in these cells. This suggests that both UPF2 and UPF3B may have specialized functions in CBs, but that UPF3A does not share those functions.
TDRD6 is necessary for assembly of chromatoid body components including UPF1 and UPF2
The exact roles of Tudor domain-containing proteins are not clear, though they seem to serve scaffolding or localization functions through interactions with DNA and RNA [48]. Fanourgakis and colleagues examined the Tudor domain-containing CB protein, TDRD6, in NMD [4]. In mice, TDRD6 exists in two isoforms, a full-length (∼250 kDa) isoform that is expressed in spermatogonia and primary spermatocytes, and a C-terminally truncated isoform (∼230 kDa) that is expressed in secondary spermatocytes and spermatids [47]. TDRD6 colocalizes with the piRNA-associated proteins DDX4, MIWI (PIWIL1), and MILI (PIWIL2) in CBs. Tdrd6−/− mice are viable, but males are infertile due to failure in production of germ cells beyond elongating spermatids. Chromatoid bodies in spermatids of Tdrd6−/− mice are diffuse and imperfectly assembled with dispersed contents, demonstrating the architectural function of TDRD6 [4]. Two interesting proteins Fanourgakis et al. identified inside intact but not TDRD6-disrupted CBs were UPF1 and UPF2, leading them to investigate roles for CBs in mechanisms of NMD.
In agreement with the hypothesis that CBs do not influence NMD of EJC-stimulated transcripts (for example, those arising from alternative splicing, Figure 1H), Fanourgakis et al. found that levels of mRNAs containing downstream exon junctions (PTC targets) did not change between Tdrd6+/− and Tdrd6−/− mice. However, as Bao et al. [2] noted with Upf2 conditional mutants, Fanourgakis et al. [4] saw in Tdrd6−/− mice an accumulation of mRNAs with long 3΄ UTRs, a hallmark of that form of NMD. This suggests that, while EJC-stimulated NMD is not a function of CBs, long 3΄ UTR-mediated NMD might be a separate pathway that is associated with the presence of CBs, and might involve UPF2 and UPF3A (see Figure 3).
Two mechanisms for nonsense-mediated decay are revealed by chromatoid bodies in male germ cells
Together, these findings suggest that there are at least two distinct mechanisms for NMD in male germ cells and, by extension, in somatic cells. One mechanism recognizes and targets mRNAs containing premature termination codons (“EJC-stimulated NMD,” most like the EJC model; Figure 2A). The second mechanism recognizes and targets mRNAs with anomalously long 3΄ UTRs (“Long 3΄ UTR NMD,” resembling the Faux 3΄ UTR model; Figure 2B). These two mechanisms might overlap with the EJC-enhanced and EJC-independent pathways described by others [49]. While both NMD mechanisms are likely active in all cells, we propose here that CBs in postmeiotic male germ cells isolate the two mechanisms such that they can act independently. It appears from the studies represented here that EJC-stimulated NMD is rare or absent in postmeiotic germ cells; neither Bao et al. [2] nor Fanourgakis et al. [4] saw evidence for it. This conclusion also agrees with earlier studies showing that NMD of specific PTC-containing transcripts was low in testis although components of NMD (Upf1, Upf2, Upf3a, and Upf3b mRNAs) were very high [50]. Meanwhile, the studies reviewed here concluded that long 3΄ UTR-mediated NMD remains active in spermatids. How might that be so?
UPF1, UPF2, UPF3A, and UPF3B are expressed in almost all cells in the body [2–4], although UPF3A is at low levels outside of male germ cells. In spermatocytes, however, UPF3B is absent and UPF3A is most prevalent (Figure 3). Similarly, in spermatids, both UPF3A and UPF3B are present, with UPF3A being generally cytoplasmic, while the majority of UPF3B and UPF2 localized to CBs. This suggests two different models to account for how long 3΄ UTR NMD is accomplished in spermatids. Neither model is perfect, but each has merit. Enterprisingly, we call these models the “Long 3΄ UTR NMD in Cytoplasm” model (Figure 4A) and the “Long 3΄ UTR NMD in CBs” model (Figure 4B).
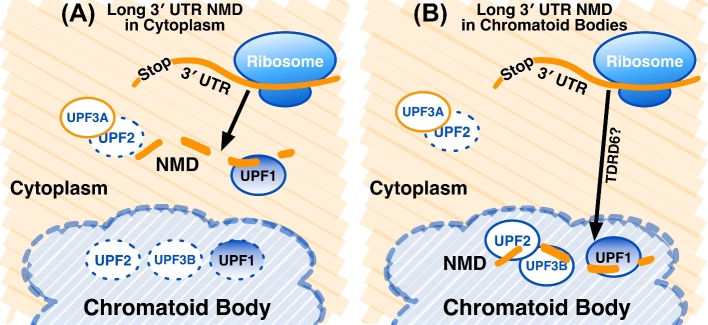
Figure 4.
Two models for the role of CBs in long 3΄ UTR-mediated NMD in spermatids. (A) The Long 3΄ UTR NMD in Cytoplasm model. UPF1, UPF2, and UPF3B are sequestered and inactive in the CB. Cytoplasmic UPF3A interferes with UPF2, turning off EJC-stimulated NMD in these cells. Long 3΄ UTR-mediated NMD occurs in the cytoplasm by normal mechanisms. (B) The Long 3΄ UTR NMD in CBs model. UPF1, UPF2, and UPF3B are localized in the CB, but remain active. Messenger RNAs with long 3΄ UTRs are identified translationally and marked for transport to the CB, possibly by TDRD6. Nonsense-mediated mRNA decay occurs in the CB.
Long 3΄ untranslated region nonsense-mediated decay in Cytoplasm model
Both models exist to explain how long 3΄ UTR NMD is active in spermatids, while EJC-stimulated NMD remains inactive. In the first model, UPF2 and UPF3B are sequestered inside CBs and thus probably not available for NMD of cytoplasmic mRNAs in CBs (Figure 4A). The majority of UPF1 appears to be in CBs as well [3], but some might remain in the cytoplasm to interact with and mark mRNA 3΄ ends. In this model, UPF3A antagonizes the activity of cytoplasmic UPF2, which would turn off EJC-stimulated NMD. Translation in the cytoplasm then targets mRNAs with long 3΄ UTRs by the Faux 3΄ UTR mechanism (Figure 2B), and degradation occurs by mechanisms involving SMG5, SMG6, or SMG7 [13]. Overall, this model agrees with known mechanisms of NMD, which do not require specialized structures in somatic cells [51]. However, it makes assumptions about the localization and roles of some of the components. For example, UPF1 is abundant in CBs [42]. UPF1, when phosphorylated, is able to bind to target 3΄ UTRs when other NMD components are less abundant [52, 53]. This suggests that UPF1 activity might be modified in ways to alter its activity in germ cells, and this might occur in CBs.
Long 3΄ untranslated region nonsense-mediated decay in chromatoid bodies model
Because the majority of UPF1, UPF2, and UPF3B in spermatids accumulate in CBs, it is attractive to hypothesize that long 3΄ UTR-mediated NMD is active inside CBs (Figure 4B). In this model, the CB milieu is permissive to long 3΄ UTR-mediated NMD, but nonpermissive to EJC-stimulated NMD. UPF3A, which is mostly cytoplasmic in spermatids, acts to turn off NMD in the cytoplasm. Perhaps TDRD6, which is necessary to localize UPF1 and UPF2 to the CBs, might also move targeted mRNAs with long 3΄ UTRs there. This model accounts for the observations that many components of NMD are localized to CBs and that disruption of CBs results in loss of long 3΄ UTR-mediated NMD [2–4]. However, a strong argument against this model is the long-standing observation that NMD requires ongoing translation to identify stop codons [34], and there is no evidence for translation in CBs (M. Wilkinson, personal communication). However, it is possible that mRNAs targeted to the CB have already been translated at least once in the cytoplasm and bear as yet undetermined marks (for example, UPF1 [54]). Studies to uncover the functions of NMD in CBs could address these opposing hypotheses.
Concluding remarks and observations
Because transcription halts in round spermatids, but translation continues into elongating spermatids, male germ cell development relies heavily on post-transcriptional modes of gene regulation in which 3΄ UTRs play prominent roles [55]. In these cells, high rates of recombination and requirements for transcript diversity result in frequent errors in mRNAs that must be surveilled and kept in check. Nonsense-mediated mRNA decay is critical, therefore, ensuring the ongoing fidelity of gene expression during germ cell development. But the idiosyncratic requirements of spermatogenesis have resulted in NMD that has been tuned to those needs. In that light, it seems that the nuances of NMD in germ cells may illuminate its principles in all cells, and we look forward to the new lessons to be learned.
Acknowledgments
The authors would like to thank Andrey Karamyshev, Charles Faust, and two anonymous reviewers for helpful comments on the manuscript. The authors also thank Miles Wilkinson and Wei Yan for sharing unpublished results.
References
- 1. Bao J, Tang C, Yuan S, Porse BT, Yan W. UPF2, a nonsense-mediated mRNA decay factor, is required for prepubertal Sertoli cell development and male fertility by ensuring fidelity of the transcriptome. Development 2015; 142:352–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bao J, Vitting-Seerup K, Waage J, Tang C, Ge Y, Porse BT, Yan W. UPF2-dependent nonsense-mediated mRNA decay pathway is essential for spermatogenesis by selectively eliminating longer 3΄ UTR transcripts. PLoS Genet 2016; 12:e1005863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shum EY, Jones SH, Shao A, Dumdie J, Krause MD, Chan WK, Lou CH, Espinoza JL, Song HW, Phan MH, Ramaiah M, Huang L et al. . The antagonistic gene paralogs Upf3a and Upf3b govern nonsense-mediated RNA decay. Cell 2016; 165:382–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fanourgakis G, Lesche M, Akpinar M, Dahl A, Jessberger R. Chromatoid body protein TDRD6 supports long 3΄ UTR triggered nonsense mediated mRNA decay. PLoS Genet 2016; 12:e1005857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev 1991; 5:2303–2314. [DOI] [PubMed] [Google Scholar]
- 6. Peltz SW, Brown AH, Jacobson A. mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev 1993; 7:1737–1754. [DOI] [PubMed] [Google Scholar]
- 7. Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci USA 2003; 100:189–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schmid R, Grellscheid SN, Ehrmann I, Dalgliesh C, Danilenko M, Paronetto MP, Pedrotti S, Grellscheid D, Dixon RJ, Sette C, Eperon IC, Elliott DJ. The splicing landscape is globally reprogrammed during male meiosis. Nucleic Acids Res 2013; 41:10170–10184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zagore LL, Grabinski SE, Sweet TJ, Hannigan MM, Sramkoski RM, Li Q, Licatalosi DD. RNA binding protein Ptbp2 is essential for male germ cell development. Mol Cell Biol 2015; 35:4030–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grozdanov PN, Amatullah A, Graber JH, MacDonald CC. TauCstF-64 mediates correct mRNA polyadenylation and splicing of activator and repressor isoforms of the cyclic AMP-responsive element modulator (CREM) in mouse testis. Biol Reprod 2016; 94:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang L, Wilkinson MF. Regulation of nonsense-mediated mRNA decay. Wiley Interdiscip Rev RNA 2012; 3:807–828. [DOI] [PubMed] [Google Scholar]
- 12. Peccarelli M, Kebaara BW. Regulation of natural mRNAs by the nonsense-mediated mRNA decay pathway. Eukaryot Cell 2014; 13:1126–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schweingruber C, Rufener SC, Zund D, Yamashita A, Muhlemann O. Nonsense-mediated mRNA decay — mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim Biophys Acta 2013; 1829:612–623. [DOI] [PubMed] [Google Scholar]
- 14. Muhlrad D, Parker R. Aberrant mRNAs with extended 3΄ UTRs are substrates for rapid degradation by mRNA surveillance. RNA 1999; 5:1299–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Toma KG, Rebbapragada I, Durand S, Lykke-Andersen J. Identification of elements in human long 3΄ UTRs that inhibit nonsense-mediated decay. RNA 2015; 21:887–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. MacDonald CC, McMahon KW. Tissue-specific mechanisms of alternative polyadenylation: testis, brain and beyond. Wiley Interdiscip Rev RNA 2010; 1:494–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lianoglou S, Garg V, Yang JL, Leslie CS, Mayr C. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev 2013; 27:2380–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weng L, Li Y, Xie X, Shi Y. Poly(A) code analyses reveal key determinants for tissue-specific mRNA alternative polyadenylation. RNA 2016; 22:813–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dass B, Tardif S, Park JY, Tian B, Weitlauf HM, Hess RA, Carnes K, Griswold MD, Small CL, MacDonald CC. Loss of polyadenylation protein τCstF-64 causes spermatogenic defects and male infertility. Proc Natl Acad Sci USA 2007; 104:20374–20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li W, Yeh HJ, Shankarling GS, Ji Z, Tian B, MacDonald CC. The τCstF-64 polyadenylation protein controls genome expression in testis. PLoS One 2012; 7:e48373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu D, Brockman JM, Dass B, Hutchins LN, Singh P, McCarrey JR, MacDonald CC, Graber JH. Systematic variation in mRNA 3΄-processing signals during mouse spermatogenesis. Nucleic Acids Res 2007; 35:234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li W, Park JY, Zheng D, Hoque M, Yehia G, Tian B. Alternative cleavage and polyadenylation in spermatogenesis connects chromatin regulation with post-transcriptional control. BMC Biol 2016; 14:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mayr C, Bartel DP. Widespread shortening of 3΄ UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 2009; 138:673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ge Z, Quek BL, Beemon KL, Hogg JR. Polypyrimidine tract binding protein 1 protects mRNAs from recognition by the nonsense-mediated mRNA decay pathway. Elife 2016; 5:e11155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu M, Hecht NB. Polypyrimidine tract binding protein 2 stabilizes phosphoglycerate kinase 2 mRNA in murine male germ cells by binding to its 3΄ UTR. Biol Reprod 2007; 76:1025–1033. [DOI] [PubMed] [Google Scholar]
- 26. Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Monch K, Thoren LA, Nielsen FC, Jacobsen SE, Nerlov C, Porse BT. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev 2008; 22:1381–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thoren LA, Norgaard GA, Weischenfeldt J, Waage J, Jakobsen JS, Damgaard I, Bergstrom FC, Blom AM, Borup R, Bisgaard HC, Porse BT. UPF2 is a critical regulator of liver development, function and regeneration. PLoS One 2010; 5:e11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Weischenfeldt J, Waage J, Tian G, Zhao J, Damgaard I, Jakobsen JS, Kristiansen K, Krogh A, Wang J, Porse BT. Mammalian tissues defective in nonsense-mediated mRNA decay display highly aberrant splicing patterns. Genome Biol 2012; 13:R35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Soumillon M, Necsulea A, Weier M, Brawand D, Zhang X, Gu H, Barthes P, Kokkinaki M, Nef S, Gnirke A, Dym M, de Massy B et al. . Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep 2013; 3:2179–2190. [DOI] [PubMed] [Google Scholar]
- 30. Gallardo T, Shirley L, John GB, Castrillon DH. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis 2007; 45:413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sadate-Ngatchou PI, Payne CJ, Dearth AT, Braun RE. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis 2008; 46:738–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol Cell Biol 2001; 21:209–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chan WK, Bhalla AD, Le Hir H, Nguyen LS, Huang L, Gecz J, Wilkinson MF. A UPF3-mediated regulatory switch that maintains RNA surveillance. Nat Struct Mol Biol 2009; 16:747–753. [DOI] [PubMed] [Google Scholar]
- 34. Jones SH, Wilkinson M. RNA decay, evolution, and the testis. RNA Biol 2017;14:146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Handel MA. The XY body: a specialized meiotic chromatin domain. Exp Cell Res 2004; 296:57–63. [DOI] [PubMed] [Google Scholar]
- 36. Yan W, McCarrey JR. Sex chromosome inactivation in the male. Epigenetics 2009; 4:452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vemuganti SA, de Villena FP, O’Brien DA. Frequent and recent retrotransposition of orthologous genes plays a role in the evolution of sperm glycolytic enzymes. BMC Genomics 2010; 11:285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tarpey PS, Raymond FL, Nguyen LS, Rodriguez J, Hackett A, Vandeleur L, Smith R, Shoubridge C, Edkins S, Stevens C, O’Meara S, Tofts C et al. . Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet 2007; 39:1127–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol 2008; 15:85–93. [DOI] [PubMed] [Google Scholar]
- 40. Schweingruber C, Soffientini P, Ruepp MD, Bachi A, Muhlemann O. Identification of Interactions in the NMD Complex Using Proximity-Dependent Biotinylation (BioID). PLoS One 2016; 11:e0150239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang P, Kang JY, Gou LT, Wang J, Xue Y, Skogerboe G, Dai P, Huang DW, Chen R, Fu XD, Liu MF, He S. MIWI and piRNA-mediated cleavage of messenger RNAs in mouse testes. Cell Res 2015; 25:193–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Meikar O, Vagin VV, Chalmel F, Sostar K, Lardenois A, Hammell M, Jin Y, Da Ros M, Wasik KA, Toppari J, Hannon GJ, Kotaja N. An atlas of chromatoid body components. RNA 2014; 20:483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Meikar O, Da Ros M, Korhonen H, Kotaja N. Chromatoid body and small RNAs in male germ cells. Reproduction 2011; 142:195–209. [DOI] [PubMed] [Google Scholar]
- 44. Kimura M, Ishida K, Kashiwabara S, Baba T. Characterization of two cytoplasmic poly(A)-binding proteins, PABPC1 and PABPC2, in mouse spermatogenic cells. Biol Reprod 2009; 80:545–554. [DOI] [PubMed] [Google Scholar]
- 45. Wang J, Saxe JP, Tanaka T, Chuma S, Lin H. Mili interacts with tudor domain-containing protein 1 in regulating spermatogenesis. Curr Biol 2009; 19:640–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yabuta Y, Ohta H, Abe T, Kurimoto K, Chuma S, Saitou M. TDRD5 is required for retrotransposon silencing, chromatoid body assembly, and spermiogenesis in mice. J Cell Biol 2011; 192:781–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vasileva A, Tiedau D, Firooznia A, Muller-Reichert T, Jessberger R. Tdrd6 is required for spermiogenesis, chromatoid body architecture, and regulation of miRNA expression. Curr Biol 2009; 19:630–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pek JW, Anand A, Kai T. Tudor domain proteins in development. Development 2012; 139:2255–2266. [DOI] [PubMed] [Google Scholar]
- 49. Metze S, Herzog VA, Ruepp MD, Muhlemann O. Comparison of EJC-enhanced and EJC-independent NMD in human cells reveals two partially redundant degradation pathways. RNA 2013; 19:1432–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zetoune AB, Fontaniere S, Magnin D, Anczukow O, Buisson M, Zhang CX, Mazoyer S. Comparison of nonsense-mediated mRNA decay efficiency in various murine tissues. BMC Genet 2008; 9:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stalder L, Muhlemann O. Processing bodies are not required for mammalian nonsense-mediated mRNA decay. RNA 2009; 15:1265–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kurosaki T, Li W, Hoque M, Popp MW, Ermolenko DN, Tian B, Maquat LE. A post-translational regulatory switch on UPF1 controls targeted mRNA degradation. Genes Dev 2014; 28:1900–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Durand S, Franks TM, Lykke-Andersen J. Hyperphosphorylation amplifies UPF1 activity to resolve stalls in nonsense-mediated mRNA decay. Nat Commun 2016; 7:12434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Boehm V, Haberman N, Ottens F, Ule J, Gehring NH. 3΄ UTR length and messenger ribonucleoprotein composition determine endocleavage efficiencies at termination codons. Cell Rep 2014; 9:555–568. [DOI] [PubMed] [Google Scholar]
- 55. Ehrmann I, Elliott DJ. Post-transcriptional control in the male germ line. Reprod Biomed Online 2005; 10:55–63. [DOI] [PubMed] [Google Scholar]