Figure 2.
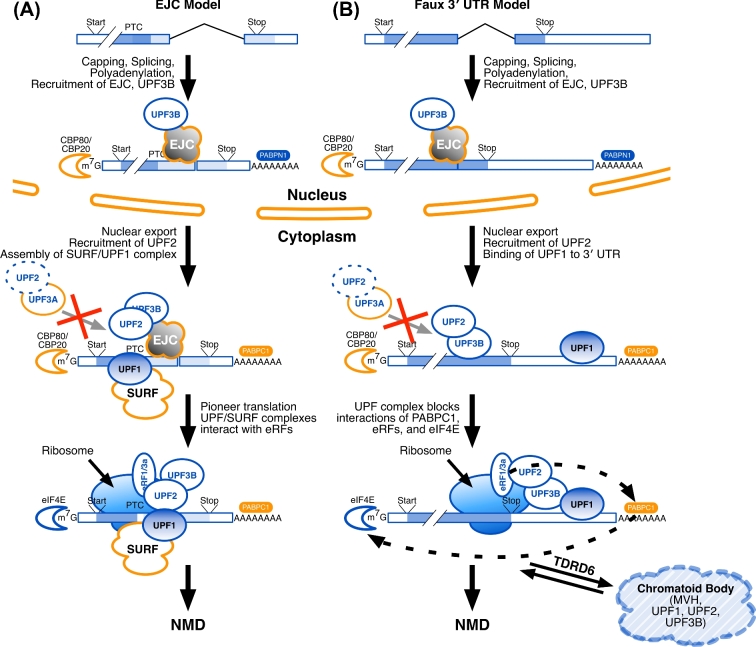
Several models vie to account for nonsense-mediated decay (NMD) in different cell types. (A) In the Exon Junction Complex (EJC) model, after splicing, EJC proteins bind at the junctions of the last and penultimate exons, to be joined by UPF3B (the EJC “marks”). This nucleoprotein complex is then exported from the nucleus to the cytoplasm, where it is joined by UPF2, the SMG1C/UPF1/eRF1/eRF3 (SURF) complex proteins, and UPF1. Nuclear poly(A) binding protein (PABPN1) is replaced by its cytoplasmic equivalent (PABPC1), and the nuclear cap-binding proteins (CBP80, CBP20) are replaced by their translational equivalents, including eIF4E. A pioneer round of translation ensues, and translational release factors (eRF1 and eRF3a) allow the ribosome to recognize the stop codon. If the stop codon is premature, that mRNA is targeted for degradation. (B) The Faux 3΄ UTR model is similar to the EJC model in its nuclear marking steps. In the cytoplasm, UPF2 binds to UPF3B. UPF1 binds to multiple RNA sites within the 3΄ UTR. In a way that is not yet understood, multiple copies of UPF1 (as might bind to a long 3΄ UTR) will interfere with an interaction between the release factors (eRF1, eRF3a) and PABPC1; multiple copies of UPF1 might also interfere with the interaction of PABPC1 and the cytoplasmic cap-binding proteins (eIF4E and others). In male germ cells, TDRD6 acts to localize UPF1, UPF2, and other factors to the CBs in postmeiotic spermatids. Also in both models (A) and (B), UPF3A may bind to UPF2 in the cytoplasm to obstruct the UPF2-UPF3B interaction and suppress EJC-stimulated NMD.