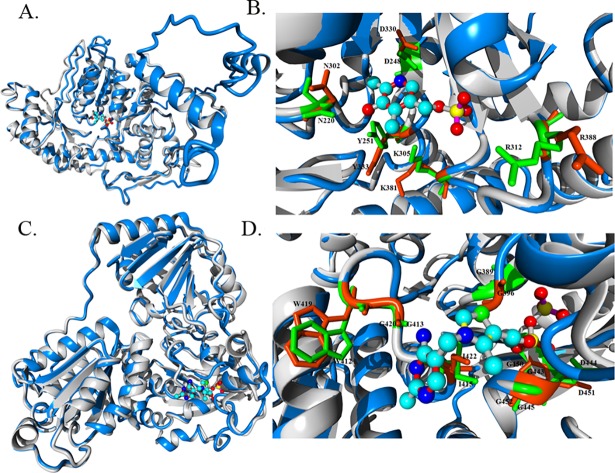
Fig 2. Structural alignment and characterization of ligand binding sites of predicted N. crassa enzymes.
Predicted structures and known templates are shown in blue and gray, respectively. Residues present in predicted enzymes and templates are indicated in orange and green, respectively. All structures are rendered as a ribbon. Key amino acid residues involved in ligand binding are rendered in a stick model. Residues in close vicinity are only highlighted. Ligands are presented in a ball and stick model. (A) The overall structural alignment of predicted ARO-8 structure with a known enzyme structure (4JE5). PLP is bound inside the enzymatic catalytic site. (B) Structural insight into the ligand binding site of both the predicted and 4JE5 known structure. (C) Structural alignment of the predicted structure of the pyruvate decarboxylase homolog from N. crassa with a known pyruvate decarboxylase structure (2VJY). TPP is bound inside the enzymatic catalytic site. (D) Characterization of the ligand binding site of the predicted enzyme using the 2VJY structure as a template.