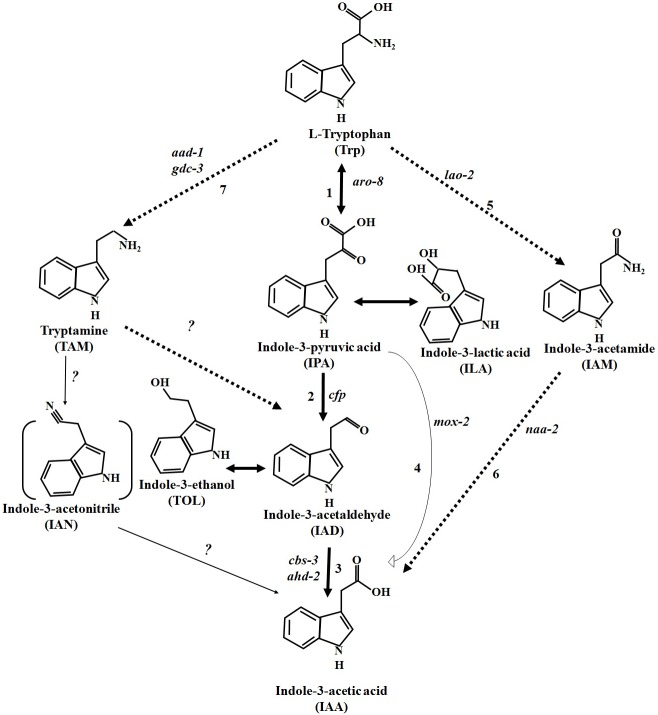
Fig 4. Proposed tryptophan-dependent IAA biosynthetic pathway in N. crassa.
The pathway with solid bold arrows was functionally characterized by the identification of novel genes, and intermediate compounds were also identified and estimated. Pathways with dashed bold arrows were proposed accordingly based on the feeding test with IAM and TAM. Solid thin arrows denote pathways proposed in green plants but not known in fungi including N. crassa. Unidentified homologues of the genes in N. crassa for the respective biosynthetic steps are indicated by question marks. IAN, with bracketed lines, has not been identified in the N. crassa culture. All other indoles without any brackets (except IAM and TAM) were identified from the culture upon tryptophan feeding. Indole-3-pyruvic acid (IPA) and indole-3-acetaldehyde (IAAld) are very unstable compounds and spontaneously convert into indole-3-lactic acid (ILA) and indole-3-ethanol (TOL), respectively. ILA and TOL can be readily identified in culture as a proxy for IPA and IAAld, respectively. Numbers in the figure reflect different enzymes: 1—tryptophan aminotransferase, 2—pyruvate decarboxylase, 3—aldehyde dehydrogenase, 4—flavin monooxygenase, 5—L-amino acid oxidase, 6—N-acylethanolamine amidohydrolase, 7 –aromatic-L-amino acid decarboxylase and glutamate decarboxylase.