Abstract
Playing a central role in cell signaling, GPCRs have evolved into the largest superfamily of membrane proteins and form the majority of drug targets in humans. How extracellular agonist binding triggers the activation of GPCRs and associated intracellular effector proteins remains, however, poorly understood. High resolution structural studies have recently revealed that inactive class-A GPCRs harbor a conserved binding site for Na+ ions in the center of their transmembrane domain, accessible from the extracellular space. Here, we show that the opening of a conserved hydrated channel in the activated state receptors allows the Na+ ion to egress from its binding site into the cytosol. Coupled with protonation changes, this ion movement occurs without significant energy barriers, and can be driven by physiological transmembrane ion and voltage gradients. We propose that Na+ ion exchange with the cytosol is a key step in GPCR activation. Further, we hypothesize that this transition locks receptors in long-lived active-state conformations.
ETOC
Vickery et al. present molecular dynamics simulations and free energy calculations, which suggest that a key step in class A GPCR activation is the exchange of a Na+ ion from an extracellular binding pocket to the cytoplasm.
INTRODUCTION
G-protein coupled receptors (GPCRs) mediate the transfer of external ligand binding information across the plasma membrane to activate a range of intracellular signaling pathways (Pierce et al. 2002). Playing a central role in regulation of vital biological systems, including nervous, cardiovascular, immune, digestive, reproductive etc., they represent the majority of membrane proteins in humans and the largest class of present drug targets (Overington et al. 2006; Rask-Andersen et al. 2014). In recent years, a number of crystal structures have been solved to reveal conformational changes between inactive and active state receptors, including common movement in transmembrane helices and conserved microswitches (Venkatakrishnan et al. 2013; Katritch et al. 2013). However, despite this wealth of structural information, it is still not fully understood how ligand binding leads to activated receptors, which are able to trigger nucleotide exchange in intracellular effector G-protein complexes.
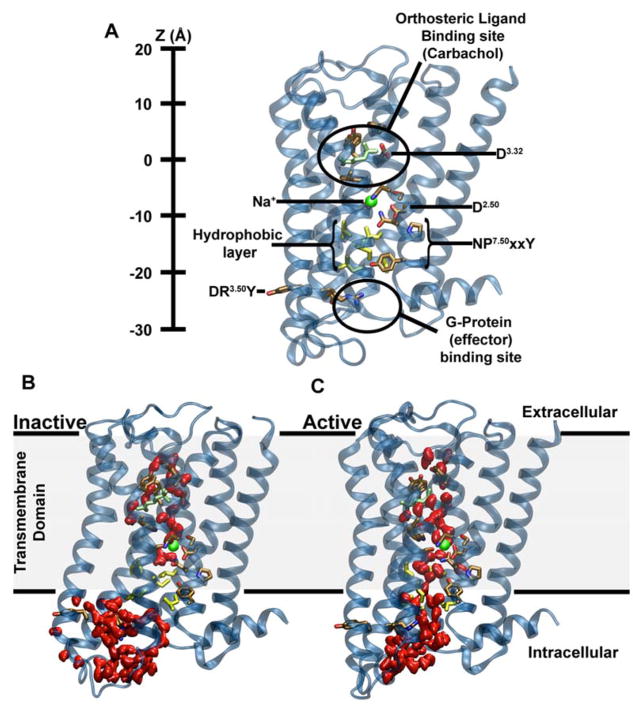
One of the major unknowns is the role of the highly conserved hydrophilic water-filled channel observed in crystal structures of class A GPCRs, which extends along the receptor axis from the external ligand-binding region nearly all the way to the effector binding site. The channel is sealed toward the cytoplasm by a thin layer of hydrophobic residues in inactive state GPCRs (Fig 1A,B). Structures of high resolution, crystallized in the inactive conformation, reveal a Na+ ion near the floor of this pocket, coordinated by water and three or four conserved residues including an acidic aspartate that is fully conserved in all ligand-sensing class A GPCRs (Fenalti et al. 2014; Zhang et al. 2012; Miller-Gallacher et al. 2014; Christopher et al. 2013; Kruse et al. 2012; Liu et al. 2012; Pardo et al. 2007) (D2.50; superscript refers to the Ballesteros and Weinstein residue numbering system) (Isberg et al. 2015). The allosteric effect of monovalent cations, in particular Na+ ions, for GPCR function has been known for almost half a century (Pert & Synder 1974), and the bulk of recent evidence shows that these effects are largely mediated by the ion binding at the D2.50 site at the physiological concentration of Na+ (140 mM and lower) (Liu et al. 2012; Massink et al. 2015; Fenalti et al. 2014). Due to the highly conserved nature of D2.50 and other Na+ ion coordinating residues, Na+ ion binding at this site is likely to be a ubiquitous feature shared by the vast majority of class A GPCRs (Katritch et al. 2014).
Figure 1. Major structural features and internal hydration of class A GPCRs in the inactive and active state as shown by the m2r.
(A) The main structural features of class A GPCRs, as exemplified by m2r, include 7 TM helices (blue), an extracellular ligand binding site, the intracellular effector (G-protein) binding site as well as conserved and functionally important residues termed microswitches (selected ones are highlighted). The vertical axis (Z-coordinate) and all positions stated in the text use the Cα atom of D1033.32 as a reference. (B) Conformation of inactive m2r (PDB: 3UON) during the simulations showing the presence of the hydrophobic layer separating the hydrophilic pocket and effector binding site. (C) After transition to the active state (PDB: 4MQT), and further simulation, m2r displays a continuous water channel connecting the orthosteric ligand binding site, hydrophilic pocket and effector binding site. Note that in (B) and (C) the upward conformation of the Y4407.53 is shown in order to highlight the changes in hydration levels only; please see Fig S3 for a detailed comparison of the upward and downward tyrosine conformations. Water molecules are shown in red (surface representation); the position of the allosteric Na+ ion, as obtained from our initial simulations, is shown as a green sphere, and residues forming the hydrophobic layer (yellow) as well as the bound ligand (carbachol, light green) are depicted in stick representation.
In active receptor conformations, the ion binding site near D2.50 shows a collapsed state, which is likely not optimal for Na+ ion binding (Liu et al. 2012; Rasmussen et al. 2011; Kruse et al. 2013; Huang et al. 2015). It was therefore proposed that the Na+ ion leaves the hydrophilic pocket upon receptor activation by a ligand or during receptor-G-protein complex formation. However, how this movement is triggered and which pathway is followed by the ion remains unknown.
Here, we investigated the link between ligand-induced receptor activation, the fate of the bound Na+ ion in class A GPCRs and its implications for transmembrane (TM) signal transduction by equilibrium and non-equilibrium atomistic simulations on the M2 muscarinic receptor (m2r). When one addresses these questions, it is important to take physiologically relevant electrochemical membrane conditions into consideration. Strong TM Na+ and K+ gradients produce a sizable voltage across the plasma membrane of up to -100 mV in the resting state of mammalian cells (Kandel et al. 2000). Both the ionic gradients and electric field have been shown to influence the function of GPCRs (Navarro-Polanco et al. 2011; Ben-Chaim et al. 2006; Rinne et al. 2015) and are likely to impact the movement of the Na+ ion within the membrane region.
Our data reveal that the Na+ ion observed in the TM domain of class A GPCRs can readily traverse the receptor and, driven by the electrochemical gradients, migrate into the cytoplasm in active receptor conformations. This result implies that a Na+ ion may be exchanged from the extracellular space to the cytoplasm as an important step in receptor activation. Furthermore, the movement of Na+ in the receptor, and intracellular egress, are coupled to a protonation change of D2.50.
RESULTS
GPCR activation opens a hydrated pathway across the receptor
We were first interested whether the conformational change from the inactive to active receptor state renders the ion-binding pocket sterically incapable of accommodating a Na+ ion. The binding site for Na+ appears to adopt a collapsed conformation in active crystal structures. We started from an inactive state structure of the m2 muscarinic acetylcholine receptor (m2r, PDB ID: 3UON) and, using a targeted molecular dynamics (TMD) approach, gently drove this conformation to the active state of this receptor (PDB ID: 4MQT) (Fig S1).
Our simulations show that the active state of m2r initially retains sufficient space for the ion. The electrostatic attraction between the ion and the negatively charged side chain of D692.50 keeps the ion bound to this site during and after the transition from the inactive to the active receptor conformation (Fig S2). However, our simulations show a widening of the intracellular portion of the TM helices below the hydrophilic pocket during this conformational change, which subsequently becomes fully hydrated (Fig 1B). The hydrated pathway forms a connection between the orthosteric ligand-binding site, the hydrophilic pocket and the G-protein binding site. The slim hydrophobic layer that delimits the hydrophilic pocket toward the G-protein binding site in the inactive crystal structure undergoes substantial conformational changes, which are especially evident from the sidechain position of Y4407.53. Our simulations show two major conformations of the Y4407.53 sidechain following the transition – an upward state similar to the conformation observed in the inactive crystal structure (PDB: 3UON; Fig S3A) and a downward configuration, which is also seen in the active crystal structure (PDB: 4MQT; Fig S3B). The formation of a hydrated pathway connecting the receptor ligand and effector binding sites has been reported in previous simulation studies on the A2AR and 5-HT1A receptors (Yuan et al. 2014; Yuan et al. 2016), however the previous reports did not take the presence of a Na+ ion into consideration.
The position of the internal Na+ ion is coupled to protonation of D2.50
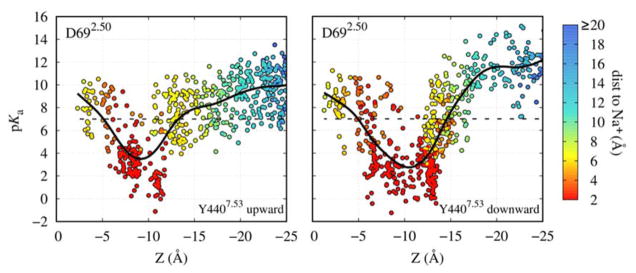
We were next interested in the interplay between the Na+ ion and the key conserved titratable residue D692.50. A number of computational studies have explored functional implications of the protonation state of D2.50, in particular its role in receptor activation, Na+ ion binding, and interaction with the “ionic lock” motif (DR3.50Y) in several class A family GPCRs (Ranganathan et al. 2014; Miao et al. 2015; Vanni et al. 2010). Here, we focused on a potential coupling between the position of the Na+ ion within the receptor and protonation of D692.50. We carried out pKa calculations on D692.50 using more than 800 equilibrated frames from simulations of the m2r receptor in a variety of conformations, including both the upward and downward configurations of the Y4407.53 sidechain. Due to the formation of a hydrated pathway across the receptor from the ligand to the effector binding sites in the active state simulations, we were able to evaluate the effect of the Na+ ion positional changes on the D692.50 pKa, where the Na+ ion was shifted both in the upward (toward the extracellular face) and downward direction.
Figure 2 shows that the pKa value and, thus, the most likely protonation state of D692.50 are substantially influenced by the Na+ ion. If the cation is within ~3–5 Å of D692.50, its positive charge strongly stabilizes the negatively charged form of D692.50, leading to a pKa value of ~3–4. However, displacement of the Na+ ion to distances of 5 Å and greater gives rise to a substantial pKa shift to values between 8–12. This can be understood given the location of D692.50 in the middle of the transmembrane domain, surrounded by many non-polar residues. Transient movements of the internal Na+ ion from its binding site, facilitated by activation-related conformational changes in the Na+ pocket, can therefore be sufficient to lead to protonation of D692.50.
Figure 2. Proximity of the Na+ ion modulates protonation of D692.50.
Continuum electrostatics calculations of the pKa of the D692.50 sidechain using a multitude of m2r conformations obtained from our atomistic simulations in the carbachol-bound active state, both for Y4407.53 in the upward (left) and downward (right) conformations. The pKa is shown as a function of Z, the separation between the Na+ ion and the Cα atom of D1033.32, which marks the orthosteric ligand binding pocket, along the TM axis (see Fig 1A). The data points are in addition coloured according to their distance to the D692.50 sidechain. The black continuous line, a smoothed spline fit, indicates the approximate average pKa for each separation for illustrative purposes, and the dashed black line shows a pKa of 7.
For the protonation of D2.50, we propose that the most likely proton entry route would be from the extracellular side, along the negative membrane potential gradient. Moreover, in the m2r and other aminergic receptors the proton could be transferred from the conserved D3.32 in the orthosteric binding pocket via a short chain of water molecules (Isom & Dohlman 2015). In the apo state, our calculations in m2r indicate that D3.32 is generally protonated (pKa = 11.2±1.7), whereas upon ligand binding the pKa is substantially lowered (pKa = 7.6±1.9). A possible protonation change of D3.32 could thus facilitate the shuttling of protons to D2.50. Furthermore, if a G-protein complex with a receptor is preformed before agonist binding, D2.50 would be readily accessible for protonation from the extracellular side via a hydrated pathway. In this context, it has further been argued that bound agonists, but not antagonists, may sustain the hydrated pathway past the ligand which connects the extracellular space with the Na+ ion binding site upon receptor activation (Yuan et al. 2016). Interestingly, the protonation state of D2.50 shows an effect on the stability of the activated receptor state in our simulations. Under equilibrium, the active state remains stable when D2.50 is neutral (Fig. S4), while it exhibits a greater propensity to revert back to the inactive state when D2.50 is charged. We obtain similar results for non-equilibrium simulations (Fig. S5). This lends further support to an important role of D2.50 protonation for receptor activation.
Simulations under electrochemical gradient show ion movement to the intracellular face
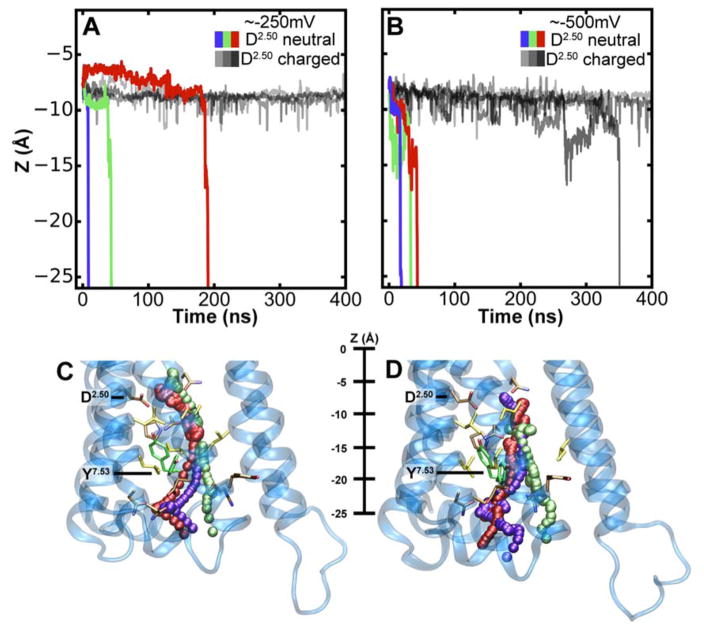
Next we conducted atomistic simulations with the Computational Electrophysiology (CompEL) protocol (Kutzner et al. 2016) on the active conformation of m2r. We applied a physiological Na+ ion gradient of 150:10 mM across the membrane from the extracellular to the intracellular side, in addition to a small ion imbalance evoking a hyperpolarised Vm at -250 mV. Due to the wide range of pKa values that D692.50 can adopt, its sidechain was modeled both in charged and neutral forms (Fig S5).
Our simulations at -250 mV show that the Na+ ion exhibits a substantial degree of mobility even when D692.50 is in the charged state (Fig 3A,B). The Na+ ion is predominantly coordinated by the residues D692.50, S1103.39, N4357.45 and S4337.46. Under a small membrane voltage, a bimodal distribution of distances between the ion and D692.50 is observed, where larger distances of 5–6 Å are not uncommon (Fig S6). As our pKa calculations showed that moderate excursions of the ion from its original binding site on this scale are likely to have a major impact on the pKa and protonation state of the D692.50 sidechain (Fig 2), we investigated the effect of a protonation change of D692.50 in the active conformation.
Figure 3. Migration of the Na+ ion across the receptor to the intracellular side.
(A–B) Z-coordinate of the Na+ ion in m2r under a hyperpolarised Vm of -250 mV (A) and -500 mV (B). Black and grey lines denote simulations with charged D692.50; purple, green and red lines display simulations with neutral D692.50. (C–D) Trajectories of the Na+ ion moving from the hydrophilic pocket, accessible from the extracellular space, into the intracellular bulk solution at -250 mV (C) and -500 mV (D). Three example trajectories are shown for each Vm; please see table S1 for a complete list. The color used to display the Na+ ion corresponds to the trajectories shown in panels A and B, respectively. Examples of the Y4407.53 upward and downward conformations are shown in green. The pathways of the ion toward the intracellular side are almost indistinguishable from each other until the ion passes Y4407.53. Thereafter, the pathways diverge to some degree due to the widened exit region to the cytoplasm.
Our simulations reveal that, in this receptor conformation, the Na+ ion readily passes through the hydrated channel into the intracellular solution. When D692.50 is neutral, we observe the Na+ ion to be expelled into the intracellular solution in three out of four simulations at -250 mV (Fig 3A,C; for a complete list of trajectories see Table S1). At -500 mV the effect is, expectably, even more pronounced and movement into the cytoplasm is seen in all four simulations we conducted (Fig 3B,D; Table S1). In contrast, when D692.50 is charged, such a transition is observed only in one out of eight simulations, namely at a raised membrane voltage (Fig 3A,B; Table S1). The observed translocation of Na+ to the intracellular side occurs irrespective of the conformation adopted by Y4407.53 (Fig 3C,D and Fig S3).
In our simulations as well as under physiological conditions, both TM ion concentration and voltage gradients drive ion flow across membrane pores. In the case of the Na+ ion, both gradients act synergistically in the resting state of the cell, driving the Na+ ion toward the cytoplasm. Under the conditions used in the simulations, fast ion motion through the receptor is predominantly voltage-driven. Converted into an effective force, and using a linear approximation to describe the gradient across the membrane (Dill & Bromberg 2011), the influence of the concentration gradient would be about 10-fold smaller (~1.3 pN) than the driving force caused by the voltage gradient under these conditions (~13 pN). At physiological conditions, both driving forces are likely to be of similar magnitude, such that ion migration could either be induced by the voltage or ion gradients.
Energetics of ion movement to the cytoplasm
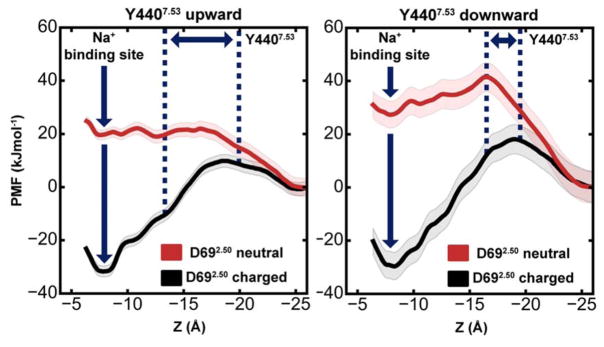
As the initiation of fast ion movement to the intracellular side was initially tested under slightly supra-physiological levels of Vm, we next evaluated the detailed equilibrium energetics of the Na+ ion movement on this pathway (i.e. without applied gradients) to ascertain the physiological relevance of this transition. We calculated the potential-of-mean-force (PMF) for the migration of the cation in four different states. In addition to probing the influence of the D692.50 protonation state, we examined the role of the conformation of the Y4407.53 sidechain, which substantially affects the width and overall shape of the formed hydrated pathway into the cytoplasm (Fig 3C,D).
When D692.50 is charged (Fig 4), the free energy difference between the internal Na+ ion binding site and the free intracellular bulk solution is ~30 kJ mol−1. Accordingly, the active conformation of m2r retains a Na+ ion at the allosteric site (Z = 7.5–8 Å) with relatively high affinity, as long as D692.50 remains deprotonated. The major barrier to migration into the cytoplasm is located near the Y4407.53 sidechain. In its upward state, the free energy barrier amounts to ~41 kJ mol−1, while it increases to ~48 kJ mol−1 in the downward state (Fig 4).
Figure 4. Energetics of Na+ translocation from the hydrophilic pocket to the intracellular side.
Equilibrium potential of mean force (PMF) profiles of the energetics of Na+ translocation along the Z-axis in m2r without any applied voltage or concentration gradients. Four relevant states were considered: (Left) negatively charged D692.50 (black) or neutral D692.50 (red) with the Y4407.53 sidechain in an upward conformation; (Right) negatively charged D692.50 (black) or neutral D692.50 (red) with a downward-oriented Y4407.53 sidechain. The standard deviation of the PMF, obtained from Bayesian bootstrap analysis, is depicted as shaded area. For each PMF, the intracellular bulk solution was used as a reference, and the range of positions adopted by the Y4407.53 sidechain is denoted by blue dotted lines.
As our pKa calculations showed that even a moderate displacement of the Na+ ion away from its binding site at D692.50 is likely to lead to a protonation change of the aspartate, we also calculated the PMF for the movement of Na+ along the intracellular pathway with neutral D692.50. Importantly, this state no longer shows any affinity for the Na+ ion, and ion movement into the intracellular bulk is not obstructed by any energy barrier significantly larger than the thermal energy (kT, ~2.5 kJ mol−1) in the upward-oriented Y4407.53 conformation. When Y4407.53 is oriented downward, a small but readily surmountable energy barrier (on physiologically relevant timescales) of ~14 kJ mol−1 exists for this transition. The downward conformation of Y4407.53, in conjunction with the neutral state of D692.50 also has a small influence on the shape and configuration of the ion binding site at D692.50, which leads to a reduction of the number of hydrogen bonds formed between the protein and the ion (Fig S7), raising the free energy of binding at this site further by ~7.5 kJ mol−1 (Fig 4A,B). In the non-equilibrium case, with a physiological Vm applied, the free energy minima at Z = ~7.5 Å will be raised with regard to the intracellular bulk by ~4.4 kJ mol−1 per 100 mV (Fig S8). This means that, in all of these states, the Na+ ion can readily traverse the receptor and permeate along a hydrated pathway to the intracellular side.
Conservation of the pocket and intracellular exit channel
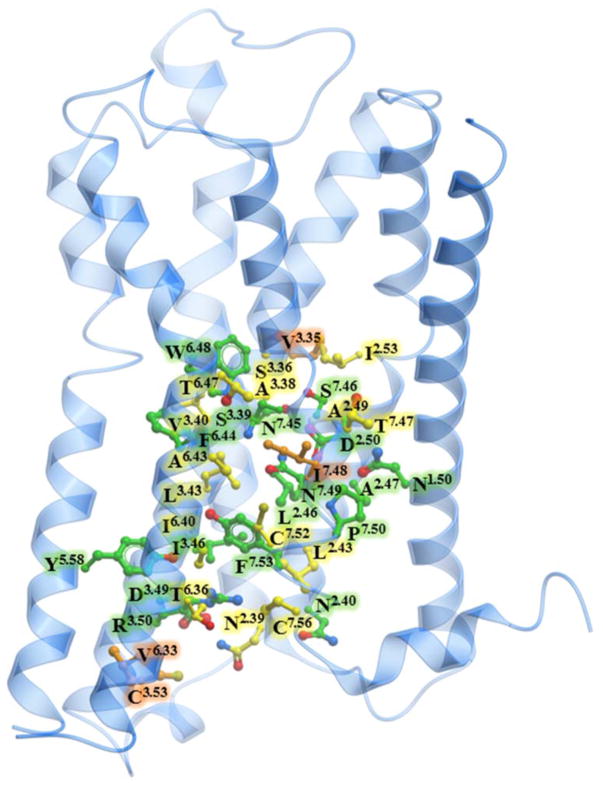
Additional support for an important role of intracellular Na+ egress in the activation of class A GPCRs is provided by an analysis of residue conservation along its exit pathway. As we detailed previously (Katritch et al. 2014), there is a remarkable level of conservation for the 16 residues of the Na+ binding pocket in class A GPCRs (Figure 5, Table S1), suggesting a conserved functional role of Na+ in receptor activation mechanism. Interestingly, our analysis of Na+ contacts along the MD trajectories in this study shows that the residues lining the ion exit path to the intracellular side are also well conserved. Thus, out of the 36 contact residues, 32 are 100% conserved among all five muscarinic receptors, 17 are >90% conserved among all aminergic receptors, and 22 are consensus residues among all class A GPCRs. Most importantly, the predicted exit pathway includes Na+ contacts with the highly conserved N1.50 (100% and 98% conserved in aminergic and in all class A respectively), D3.49 (100% and 64%), Y5.58 (94% and 73%) and other residue positions generally conserved as polar residues, including N1.60, T2.37 and N2.39. Particularly, in the inactive M2 muscarinic receptor and in other inactive state GPCR structures as well, the Y7.53 residue is directed toward the Na+ ion-binding pocket, and hence may play a role as first point of polar contact outside the Na+ ion-binding pocket for the intracellular movement of Na+. Na+ ion passage toward the cytosol may be further facilitated by other conserved polar residues, including D3.49, N2.39, N2.40 and T2.37. The conservation of the Na+ ion pocket and the path for intracellular egress of Na+ suggests that the Na+ transfer described in this study can occur in all muscarinic receptors and other class A GPCRs, comprising a key “irreversible” part of the activation mechanism.
Figure 5. Conservation of the intracellular Na+ ion pathway.
Muscarinic m2 receptor shown in blue cartoon representation, along with ball-and-stick representation of residues involved in the egress of the Na+ ion. The carbon atoms of 17 residues that are >90% conserved among aminergic receptors are shown in green, the carbon atoms of additional 15 residues that are conserved among the muscarinic family of receptors are shown in yellow, the carbon atoms of the 4 non-conserved residues are shown in orange.
DISCUSSION
The principal role of GPCRs is to transmit information about an extracellular agonist binding event toward the cytoplasm, by catalyzing GDP release from a bound intracellular G-protein complex (Pierce et al. 2002). This is known to involve conformational changes in the receptor, including conserved residue microswitches, and large scale movement of TM helices 6 and 7 on the intracellular side that open the nucleotide binding site of the Gα protein (Mahoney & Sunahara 2016; Dror et al. 2015; Huang et al. 2015). It has, furthermore, long been recognized that G-protein binding, and stabilization of this conformation on the intracellular side of the receptor, increases agonist affinity on the extracellular face (Maguire et al. 1976; DeVree et al. 2016).
Na+ ions, binding to an internal receptor site between the G-protein and the external ligand binding pockets, are known to act as powerful allosteric modulators of class A GPCRs (Pert & Synder 1974; Katritch et al. 2014). Na+ was found to selectively diminish the affinity of agonists, but not antagonists, to GPCRs, which can be interpreted as a structural stabilization of the inactive receptor state by the ions (Miller-Gallacher et al. 2014; Selley et al. 2000; Quitterer et al. 1996). Accordingly, while receptor X-ray structures of sufficient resolution crystallized in the inactive state display a Na+ ion bound to D2.50, this binding site is collapsed in active receptor conformations, and ions are not observed (Huang et al. 2015; Katritch et al. 2014). Mutations around the Na+ ion binding site have a major impact on receptor function in most class A GPCRs (Liu et al. 2012), either completely abolishing G-protein activation, or resulting in constitutive ligand-independent or pathway-biased signaling (Liu et al. 2012; Massink et al. 2015; Fenalti et al. 2014).
Our work shows that the Na+ ion binding pocket, which is accessible only from the extracellular face in the inactive state (Selent et al. 2010; Vickery et al. 2016), is transformed into a fully permeable, water-filled channel in the activated receptor conformation of m2r (Fig S9). This channel bridges the extracellular ligand and intracellular G-protein binding sites. Water access from the ligand binding site all the way to the cytoplasmic side of the receptor has previously also been observed in simulations on the A2AR and 5-HT1A receptors (Yuan et al. 2014; Yuan et al. 2016). We show here that the activated receptor state permits the Na+ ion to readily cross the receptor toward the cytoplasmic side without experiencing major energy barriers on its pathway. The high hydration level of this pathway in the active state is thereby an important factor in facilitating ion passage. A correlation between hydration level and ion transfer has previously been demonstrated in the case of ion channels (Dong et al. 2013; Zhu & Hummer 2012; Beckstein et al. 2003). In simulations of the inactive state, by contrast, the application of substantially larger forces seems to be necessary to achieve inward migration of Na+, as no continuous hydrated channel is formed (Shang et al. 2014).
The inward motion of the Na+ ion is likely facilitated by a protonation change of D2.50 from the negatively charged to the neutral form, which we show to occur even upon small displacements of the ion from its equilibrium binding position. Neutralization of D2.50 substantially reduces the affinity of the binding site for Na+ ions. Migration of the ion toward the cytosol is then driven by the negative membrane voltage and by a greater than 10-fold Na+ gradient across the cytoplasmic membrane under physiological conditions, both strongly attracting Na+ ions inward. Indeed, we observe that moderately negative membrane voltages allow fast escape of the allosteric Na+ ion to the cytoplasm on 10–100 ns-timescales in our simulations.
According to our results, conformational changes associated with agonist binding from the extracellular side and/or G-protein binding from the cytoplasm alter the Na+ site conformation and the dynamics of the Na+–D2.50 pair. This, in turn, leads to a protonation change of this residue, and subsequent egress of the Na+ ion via a hydrated exit channel to the intracellular side.
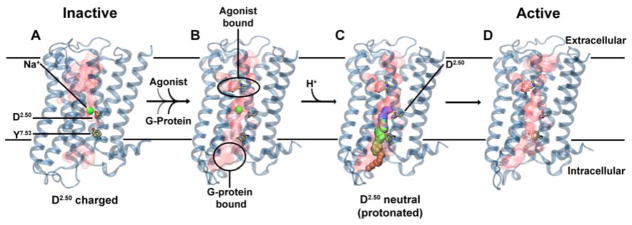
We therefore suggest that intracellular Na+ ion transfer, facilitated by the membrane potential and Na+ gradient, is a pivotal step during receptor activation. We further hypothesize that this transition traps the receptor in the active state (Fig 6). The loss of Na+ is associated with receptor activation, and it has been shown that, once activated, GPCRs remain in a prolonged active state, capable of signaling even when the receptors are internalized from the cytoplasmic membrane during endocytosis (Thomsen et al. 2016; Irannejad et al. 2013). The crucial role of the Na+ ion movement within the receptor is reflected by the nearly complete conservation of the Na+ ion binding site in class A GPCRs, as well as the high conservation level of the exit pathway. The mechanism suggested here is also consistent with agonist independent basal signaling of GPCRs (Kobilka & Deupi 2007), explaining this phenomenon as spontaneous protonation of D2.50 and egress of the bound Na+ ion on the intracellular pathway, leading to receptor activation. Following arrival on the cytoplasmic side, it is conceivable that the ion induces further conformational transitions through its strong interaction with protein residues, including at the G-protein-receptor interface and the G-protein itself. This region includes a number of charged and polar groups, for example a polar network extending across all G-proteins, similar to the one observed in GPCRs which enables ion movement (Isom & Dohlman 2015; Isom et al. 2013).
Figure 6. Proposed role of Na+ translocation in GPCR activation.
Key checkpoints during the transition from the inactive (A) to active (D) state of the receptor. (A) The initial, inactive receptor conformation shows no bound agonist or G-protein, and displays a Na+ ion bound in a pocket which is sealed towards the cytosol by a hydrophobic layer around Y7.53. (B) G-protein and agonist bind to the receptor (in undetermined order), leading to the formation of a continuous water channel across the GPCR. The increased mobility of the Na+ ion results in a pKa shift and subsequent protonation of D2.50. (C) Neutralization of D2.50 and the presence of the hydrated pathway facilitate transfer of Na+ to the intracellular side, driven by the transmembrane Na+ gradient and the negative cytoplasmic membrane voltage. (D) The expulsion of Na+ towards the cytosol results in a prolonged active state of the receptor.
Charge movements within membrane proteins, such as the coupled transfer of Na+ ions and protons suggested by our MD simulations and pKa calculations, should be sensitive to the membrane voltage. Indeed, it has been demonstrated that GPCR signaling is modulated by membrane voltage changes (Vickery, Machtens, Tamburrino, et al. 2016; Martinez-Pinna et al. 2004; Rinne et al. 2015; Mahaut-Smith et al. 2008; Ben-Chaim et al. 2006; Moreno-Galindo et al. 2016). This applies both to the conformation of the receptors as well as their transmitted signal. Our findings are therefore consistent with these observations, as they suggest that movement of ions in the receptors constitute a key element in the receptor activation process. The observed voltage regulation of GPCRs is of particular relevance for receptors expressed in electrically excitable cells (Heifetz et al. 2016). In these cell types, the membrane voltage undergoes large-scale oscillations during action potentials. The transmitted receptor signal could thereby be tuned depending on the specific cell type and its excitation status (Vickery, 2016). Crucially, many GPCR drug targets are located in excitable tissue in the brain or muscle, where voltage regulation and a differential response to drugs may play an important role.
To summarize, our results suggest a model for class A GPCR activation, in which conformational changes induced by G-protein and agonist binding are accompanied by the intracellular transfer of an internally bound Na+ ion. Importantly, these conformational changes encompass rearrangement of the sidechain of Y7.53, a conserved receptor microswitch (Katritch et al. 2013), which in its upward state allows nearly barrier-free intracellular permeation of Na+ ions. This observation forms a functional link between the major Na+ binding site D2.50 and Y7.53 as the first polar point of contact on the intracellular migration pathway of the Na+ ion. Translocation of the ion is facilitated by protonation of the conserved D2.50 residue (Fig 6) and driven by the physiological membrane Na+ and voltage gradients. The voltage sensitivity of GPCRs, which has been previously reported for many receptors (Vickery, 2016), would thus be a natural consequence of an activation mechanism which incorporates the movement of ions as a key element. The Na+ free receptors are likely to be trapped in an active state, potentially explaining the prolonged mechanisms of signaling observed in many GPCRs. Our results suggest a link between TM signal transduction by receptor proteins and the voltage and ion-gradient driven permeation of ions across ion channels and pores, forming the basis of electric signal transduction in cells.
Based on our findings, we further speculate that the ligand-induced translocation of an ion across the receptor may reflect a common functional principle, which links microbial 7-transmembrane proteins with the structurally remarkably similar eukaryotic GPCRs. The function of microbial 7-transmembrane proteins, such as bacteriorhodopsin and channelrhodopsins, is to transport protons and ions across the membrane following the absorption of photons (Ernst et al. 2014; Mirzadegan & Benko 2003).
EXPERIMENTAL PROCEDURES
The simulation system for the m2r in the inactive state was constructed using the crystal structure (PDB: 3UON)(Haga et al. 2012). Ligands and non-GPCR subunits were removed. The missing loop ICL3 was modelled using Modeller (v9.14)(Šali & Blundell 1993). All internal water molecules and ions were retained, and a Na+ ion was positioned into the hydrophilic pocket. The charged N- and C-termini were capped using acetyl and methyl moieties, respectively. All ionisable groups were simulated with default protonation states, unless otherwise mentioned. The receptor was embedded into an equilibrated and hydrated 1,2-palmitoyl-oleoyl-sn-glycero-3-phosphocholine (POPC) lipid bilayer using the GROMACS utility g_membed (Wolf et al. 2010) resulting in a system size of ~92 × 88 × 97 Å. A concentration of 150 mM NaCl in the aqueous solution was used for the single bilayer systems. During equilibration, all protein heavy atoms were position-restrained with a force constant of 1000 kJ mol−1 nm−2 for 5–10 ns. Due to the low degree of internal hydration and medium resolution of the m2r structure, the equilibration was extended by another 100 ns, now without position restraints, to enable full hydration of the hydrophilic pocket.
To study the active structure, the ligand carbachol was parameterised using AMBER16, GAFF2, AM1-BCC parameters (Case et al. 2016), and docked into the orthosteric ligand binding site using GOLD (v5.2.2). We then used a targeted MD (TMD) approach with the RMSD to the protein Cα atoms of the active m2r crystal structure (PDB: 4MQT) as a reference, in order to gently enforce the transition from the inactive (PDB: 3UON) to the active state, and further equilibrated it for ~250 ns. While the backbone rapidly transitioned toward the active conformation (Fig. S1), the adaptation of sidechains and the increase in hydration of the receptor occurred on a slightly slower timescale, necessitating this simulation time span. The two major conformations of Y4407.53 observed during this simulation were then probed systematically in the PMF calculations, in which distance restraints between N1.50-Cα and D2.50-Cα to Y7.53-Cζ and dihedral restraints on the sidechain of Y7.53 were used to maintain the protein in the conformations of interest. To keep the G-protein binding site in an active conformation despite the absence of bound G-protein, we applied, at this interaction site, a minimal set of four distance restraints to the Cα atoms of the terminal groups of TM helices 2, 5, 6 and 7, namely between residues 2.39–6.33, 2.39–5.61, 2.43–7.54 and 6.36–7.54 (Fig S10).
For the CompEL simulations, the aforementioned active system was duplicated along the Z axis to construct double bilayer systems. A NaCl gradient of 150mM:10mM between the extracellular and intracellular compartments was used, along with an ion imbalance of 1 to 2 Cl− ions to generate a Vm of ~-250 to ~-500 mV, as previously described (Kutzner et al. 2011). The Vm was determined by the GROMACS utility gmx potential.
To calculate the PMF for Na+ ion translocation across m2r at neutral Vm, umbrella sampling calculations were performed in bins of 0.25 Å and analysed with the GROMACS utility gmx wham. We used a simulation time of 50 ns in each window and harmonic potentials of 900–2000 kJ mol−1 nm−2 to restrain the Na+ ion in the Z-direction. The standard deviation of the PMF profiles was estimated by using the Bayesian bootstrap method, as implemented in gmx wham, with 200 runs. The free energy of the Na+ ion in bulk solution was set to 0. The position of the Na+ ion (Z-coordinate) is reported relative to the D1033.32-Cα atom (ligand binding site).
To calculate the gating charges, we followed a method previously described in Vickery et al., 2016 and Machtens et al., 2017. A single bilayer of the active system was duplicated along the Z-axis, with one bilayer inverted (intracellular components of the receptors facing each other). The charge imbalance between compartments was then neutralised by adding ions. All protein atoms except hydrogen atoms were position-restrained using a spring constant of 100 kJ mol−1 nm−2, whilst the Na+ ion was restrained with a force constant of 10,000 kJ mol−1 nm−2 due to its greater mobility. Bulk Na+ ions were position restrained on the Z-axis using a spring constant of 200 kJ mol−1 nm−2 to prevent their ingress into the receptor. The systems were calibrated using charge imbalances of -4 to 4; the slopes of the charge imbalance-voltage relationships indicate a near constant capacitance of the membrane/protein system under these conditions. The gating charges were then inferred from the voltage differences for each ion position at a given charge imbalance. The errors were derived from the maximum and minimum slopes of the charge imbalance-voltage relationships. The hydrophilic channel was scanned, by placing the ion at 2.5 Å intervals from the hydrophilic pocket to the intracellular solution and simulated for 50 ns, with the first 5 ns discarded (Fig S8 E). The gating charge calculated for each interval was taken as a direct measure of the voltage drop within the hydrated channel. This voltage drop, multiplied by the elementary charge e for a monovalent ion, was added to the equilibrium PMFs obtained by umbrella sampling (Fig S8 A–D), representing the excess free energy.
For all MD simulations, the amber99sb_ildn force field was used for the protein (Lindorff-Larsen et al. 2010), Berger parameters for lipids (Berger et al. 1997), which were adapted for use with the amber99sb force field (Cordomí et al. 2012), and the SPC/E model for water molecules (Berendsen et al. 1987). Water bond angles and distances were constrained by SETTLE (Miyamoto & Kollman 1992) while all other bonds were constrained using the LINCS method (Hess et al. 1997). The temperature and pressure were kept constant throughout the simulations at 310 K and 1 bar, respectively, with the protein, lipids, and water/ions coupled individually to a temperature bath by the v-rescale method using a time constant of 0.2 ps and a semi-isotropic Berendsen barostat (Bussi et al. 2007; Berendsen et al. 1984). Employing a virtual site model for hydrogen atoms (Feenstra et al. 1999) allowed the use of 4-fs time steps during the simulation. All simulations were performed with the GROMACS software package, version 5.1.2 (Abraham et al. 2015).
The pKa calculations were performed using a continuum electrostatics method, namely the Poisson-Boltzmann/Monte Carlo (PB/MC) approach, on multiple snapshots taken at a 2 ns interval from different umbrella sampling simulations. PB calculations were performed using MEAD (version 2.2.9)(Bashford & Gerwert 1992) with a dielectric constant (εp) of 4 for the protein and 80 for the solvent (εw), in the presence of an explicit membrane. The temperature was set to 310 K and the ionic strength to 0.145 M. The same temperature was used for MC calculations (103 steps in each calculation), which were performed using MCRP (Baptista et al. 1999). Each MC step consisted of a cycle of random choices of a state for all individual sites and pairs of sites with couplings above 2.0 pKa units (Baptista et al. 1999), whose acceptance/rejection followed a Metropolis criterion (Metropolis et al. 1953); tautomeric forms were not included.
The GROMACS software package, version 5.0.4 analysis toolkit was used to identify residues with non-hydrogen heavy atoms within 4 Å of the sodium ion path during the simulations. The residue conservation profile of the amino acids was obtained from the GPCRdb server (Isberg et al. 2015).
Supplementary Material
HIGHLIGHTS.
Class A GPCRs possess a conserved internal binding site for Na+ ions.
Active receptors show barrier-free permeation of the Na+ ion into the cytoplasm.
Ion permeation is driven by membrane voltage and Na+ gradients.
Our results suggest that Na+ ion exchange forms a crucial step in GPCR activation.
Acknowledgments
This work was supported by the BBSRC (Training Grant BB/J013072/1 to U.Z.) and the Scottish Universities’ Physics Alliance (C.A.C, A.V.P. and U.Z.). This research was partially supported by National Institute of Health grant DA035764 to V.K. We thank Salomé Llabrés and Daniel Seeliger for fruitful discussions.
Footnotes
CONTRIBUTIONS
Conceptualization, V.K., U.Z; Methodology, O.N.V, C.A.C, S.A.Z, A.V.P, V.K, U.Z; Analysis, O.N.V, C.A.C, S.A.Z, A.V.P, V.K, U.Z; Investigation, O.N.V, C.A.C, S.A.Z; Writing – Original draft O.N.V, C.A.C, A.V.P, V.K and U.Z; Writing – Review and editing, O.N.V, C.A.C, S.A.Z, A.V.P, V.K, U.Z; Funding acquisition, V.K. and U.Z; Supervision, V.K. and U.Z.
References
- Abraham MJ, et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. Available at: http://linkinghub.elsevier.com/retrieve/pii/S2352711015000059. [Google Scholar]
- Baptista AM, Martel PJ, Soares CM. Simulation of Electron-Proton Coupling with a Monte Carlo Method: Application to Cytochrome c3 Using Continuum Electrostatics. Biophysical Journal. 1999;76(6):2978–2998. doi: 10.1016/S0006-3495(99)77452-7. Available at: http://linkinghub.elsevier.com/retrieve/pii/S0006349599774527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashford D, Gerwert K. Electrostatic calculations of the pKa values of ionizable groups in bacteriorhodopsin. Journal of Molecular Biology. 1992;224(2):473–486. doi: 10.1016/0022-2836(92)91009-e. Available at: http://linkinghub.elsevier.com/retrieve/pii/002228369291009E. [DOI] [PubMed] [Google Scholar]
- Beckstein O, et al. Ion channel gating: Insights via molecular simulations. FEBS Letters. 2003;555(1):85–90. doi: 10.1016/s0014-5793(03)01151-7. [DOI] [PubMed] [Google Scholar]
- Ben-Chaim Y, et al. Movement of “gating charge” is coupled to ligand binding in a G-protein-coupled receptor. [Accessed March 19, 2014];Nature. 2006 444(7115):106–9. doi: 10.1038/nature05259. Available at: http://www.ncbi.nlm.nih.gov/pubmed/17065983. [DOI] [PubMed] [Google Scholar]
- Berendsen HJC, et al. Molecular dynamics with coupling to an external bath. The Journal of Chemical Physics. 1984;81:3684–3690. Available at: http://link.aip.org/link/JCPSA6/v81/i8/p3684/s1&Agg=doi%5Cnpapers2://publication/doi/10.1063/1.448118. [Google Scholar]
- Berendsen HJC, Grigera JR, Straatsma TP. The Missing Term in Effective Pair Potentials. Journal of Physical Chemistry. 1987;91(24):6269–6271. Available at: http://links.isiglobalnet2.com/gateway/Gateway.cgi?GWVersion=2&SrcAuth=mekentosj&SrcApp=Papers&DestLinkType=FullRecord&DestApp=WOS&KeyUT=A1987K994100038%5Cnpapers2://publication/uuid/17978EF7-93C9-4CB5-89B3-086E5D2B9169%5Cnhttp://pubs.acs.org/doi/pdf/10.1021/ [Google Scholar]
- Berger O, Edholm O, Jähnig F. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophysical journal. 1997 May;72:2002–2013. doi: 10.1016/S0006-3495(97)78845-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. Journal of Chemical Physics. 2007:126. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- Case D, et al. Amber 2016 2016 [Google Scholar]
- Christopher JA, et al. Biophysical Fragment Screening of the β 1 -Adrenergic Receptor: Identification of High Affinity Arylpiperazine Leads Using Structure-Based Drug Design. Journal of Medicinal Chemistry. 2013;56(9):3446–3455. doi: 10.1021/jm400140q. Available at: http://pubs.acs.org/doi/abs/10.1021/jm400140q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordomí A, Caltabiano G, Pardo L. Membrane protein simulations using AMBER force field and Berger lipid parameters. Journal of Chemical Theory and Computation. 2012;8:948–958. doi: 10.1021/ct200491c. [DOI] [PubMed] [Google Scholar]
- DeVree BT, et al. Allosteric coupling from G protein to the agonist-binding pocket in GPCRs. Nature. 2016;535(7610):182–6. doi: 10.1038/nature18324. Available at: http://www.nature.com/doifinder/10.1038/nature18324%5Cnhttp://www.ncbi.nlm.nih.gov/pubmed/27362234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill KA, Bromberg S. Molecular Driving Force. 2011 Available at: papers2://publication/uuid/0936960F-D323-459D-87B8-37DE4510402C.
- Dong H, et al. Pore waters regulate ion permeation in a calcium release-activated calcium channel. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:17332–7. doi: 10.1073/pnas.1316969110. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3808583&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror RO, et al. Structural basis for nucleotide exchange in heterotrimeric G proteins. Science. 2015;348(6241):1361–1365. doi: 10.1126/science.aaa5264. Available at: http://www.sciencemag.org/cgi/doi/10.1126/science.aaa5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst OP, et al. Microbial and animal rhodopsins: Structures, functions, and molecular mechanisms. Chemical Reviews. 2014;114(1):126–163. doi: 10.1021/cr4003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feenstra KA, Hess B, Berendsen HJC. Improving efficiency of large time-scale molecular dynamics simulations of hydrogen-rich systems. Journal of Computational Chemistry. 1999;20(8):786–798. doi: 10.1002/(SICI)1096-987X(199906)20:8<786::AID-JCC5>3.0.CO;2-B. Available at: http://dx.doi.org/10.1002/(SICI)1096-987X(199906)20:8%3C786::AID-JCC5%3E3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Fenalti G, et al. Molecular control of δ-opioid receptor signalling. [Accessed March 21, 2014];Nature. 2014 506(7487):191–196. doi: 10.1038/nature12944. Available at: http://www.ncbi.nlm.nih.gov/pubmed/24413399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga K, et al. Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature. 2012;482(7386):547–551. doi: 10.1038/nature10753. Available at: http://dx.doi.org/10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heifetz A, et al. Guiding lead optimization with GPCR structure modeling and molecular dynamics. Current Opinion in Pharmacology. 2016;30:14–21. doi: 10.1016/j.coph.2016.06.004. Available at: http://dx.doi.org/10.1016/j.coph.2016.06.004. [DOI] [PubMed] [Google Scholar]
- Hess B, et al. LINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry. 1997;18(12):1463–1472. Available at: http://doi.wiley.com/10.1002/(SICI)1096-987X(199709)18:12%3C1463::AID-JCC4%3E3.0.CO;2-H. [Google Scholar]
- Huang W, et al. Structural insights into μ-opioid receptor activation. Nature. 2015;524(7565):315–321. doi: 10.1038/nature14886. Available at: http://www.nature.com/doifinder/10.1038/nature14886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irannejad R, et al. Conformational biosensors reveal GPCR signalling from endosomes. Nature. 2013;495(7442):534–538. doi: 10.1038/nature12000. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3835555&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isberg V, et al. Generic GPCR residue numbers – aligning topology maps while minding the gaps. Trends in Pharmacological Sciences. 2015;36(1):22–31. doi: 10.1016/j.tips.2014.11.001. Available at: http://dx.doi.org/10.1016/j.tips.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isom D, et al. Protons as second messenger regulators of G protein signaling. Molecular Cell. 2013;51(4):531–538. doi: 10.1016/j.molcel.2013.07.012. Available at: http://dx.doi.org/10.1016/j.molcel.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isom DG, Dohlman HG. Buried ionizable networks are an ancient hallmark of G protein-coupled receptor activation. Proceedings of the National Academy of Sciences. 2015;2015:201417888. doi: 10.1073/pnas.1417888112. Available at: http://www.pnas.org/lookup/doi/10.1073/pnas.1417888112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ER, Schwartz JH, Jessell TM. Principles of Neural Science 2000 [Google Scholar]
- Katritch V, et al. Allosteric sodium in class A GPCR signaling. [Accessed May 27, 2014];Trends in Biochemical Sciences. 2014 39(5):233–244. doi: 10.1016/j.tibs.2014.03.002. Available at: http://www.ncbi.nlm.nih.gov/pubmed/24767681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katritch V, Cherezov V, Stevens RC. Structure-function of the G protein-coupled receptor superfamily. [Accessed April 30, 2014];Annual review of pharmacology and toxicology. 2013 53:531–56. doi: 10.1146/annurev-pharmtox-032112-135923. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3540149&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobilka BK, Deupi X. Conformational complexity of G-protein-coupled receptors. Trends in Pharmacological Sciences. 2007;28(8):397–406. doi: 10.1016/j.tips.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Kruse AC, et al. Activation and allosteric modulation of a muscarinic acetylcholine receptor. [Accessed May 2, 2014];Nature. 2013 504(7478):101–106. doi: 10.1038/nature12735. Available at: http://www.nature.com/doifinder/10.1038/nature12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse AC, et al. Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature. 2012;482:552–556. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutzner C, et al. Biochimica et Biophysica Acta Insights into the function of ion channels by computational electrophysiology simulations. BBA - Biomembranes. 2016 doi: 10.1016/j.bbamem.2016.02.006. Available at: http://dx.doi.org/10.1016/j.bbamem.2016.02.006. [DOI] [PubMed]
- Kutzner C, et al. Computational electrophysiology: the molecular dynamics of ion channel permeation and selectivity in atomistic detail. [Accessed July 14, 2014];Biophysical journal. 2011 101(4):809–17. doi: 10.1016/j.bpj.2011.06.010. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3175076&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindorff-Larsen K, et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. [Accessed July 17, 2014];Proteins. 2010 78(8):1950–8. doi: 10.1002/prot.22711. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2970904&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, et al. Structural Basis for Allosteric Regulation of GPCRs by Sodium Ions. [Accessed May 1, 2014];Science. 2012 337(6091):232–236. doi: 10.1126/science.1219218. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3399762&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machtens JP, Briones R, Alleva C, de Groot BL, Fahlke C. Gating Charge Calculations by Computational Electrophysiology Simulations. Biophysical Journal. 2017;112(7):1396–1405. doi: 10.1016/j.bpj.2017.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire ME, Van Arsdale PM, Gilman aG. An agonist-specific effect of guanine nucleotides on binding to the beta adrenergic receptor. Molecular pharmacology. 1976;12:335–339. [PubMed] [Google Scholar]
- Mahaut-Smith MP, Martinez-Pinna J, Gurung IS. A role for membrane potential in regulating GPCRs? [Accessed November 12, 2014];Trends in Pharmacological Sciences. 2008 29(8):421–429. doi: 10.1016/j.tips.2008.05.007. Available at: http://www.ncbi.nlm.nih.gov/pubmed/18621424. [DOI] [PubMed] [Google Scholar]
- Mahoney JP, Sunahara RK. Mechanistic insights into GPCR-G protein interactions. Current opinion in structural biology. 2016;41:247–254. doi: 10.1016/j.sbi.2016.11.005. Available at: http://dx.doi.org/10.1016/j.sbi.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Pinna J, et al. Sensitivity limits for voltage control of P2Y receptor-evoked Ca2+ mobilization in the rat megakaryocyte. [Accessed November 18, 2014];The Journal of physiology. 2004 555(Pt 1):61–70. doi: 10.1113/jphysiol.2003.056846. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1664815&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massink A, et al. Sodium Ion Binding Pocket Mutations and Adenosine A2A Receptor Function. Molecular Pharmacology. 2015;87(2):305–313. doi: 10.1124/mol.114.095737. Available at: http://molpharm.aspetjournals.org/cgi/doi/10.1124/mol.114.095737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metropolis N, et al. Equation of state calculations by fast computing machines. Journal Chemical Physics. 1953;21(6):1087–1092. Available at: http://jcp.aip.org/resource/1/jcpsa6/v21/i6/p1087_s1?bypassSSO=1. [Google Scholar]
- Miao Y, Caliman AD, McCammon JA. Allosteric Effects of Sodium Ion Binding on Activation of the M3 Muscarinic G-Protein-Coupled Receptor. Biophysical Journal. 2015;108(7):1796–1806. doi: 10.1016/j.bpj.2015.03.003. Available at: http://dx.doi.org/10.1016/j.bpj.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Gallacher JL, et al. The 2.1 Å Resolution Structure of Cyanopindolol-Bound β1-Adrenoceptor Identifies an Intramembrane Na+ Ion that Stabilises the Ligand-Free Receptor. [Accessed March 26, 2014];PloS one. 2014 9(3):e92727. doi: 10.1371/journal.pone.0092727. Available at: http://www.ncbi.nlm.nih.gov/pubmed/24663151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzadegan T, Benko G. Sequence Analyses of G-Protein-Coupled Receptors: Simi- larities to Rhodopsin - Corrections. Biochemistry. 2003;42(10):2759–2767. doi: 10.1021/bi027224+. Available at: http://pubs3.acs.org/acs/journals/doilookup?in_doi=10.1021/bi027224+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S, Kollman PA. SETTLE: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. Journal of computational chemistry. 1992;13:952–962. Available at: http://onlinelibrary.wiley.com/doi/10.1002/jcc.540130805/abstract. [Google Scholar]
- Moreno-Galindo EG, et al. The agonist-specific voltage dependence of M2 muscarinic receptors modulates the deactivation of the acetylcholine-gated K+ current (I KACh) Pflügers Archiv - European Journal of Physiology. 2016 doi: 10.1007/s00424-016-1812-y. Available at: http://link.springer.com/10.1007/s00424-016-1812-y. [DOI] [PubMed]
- Navarro-Polanco Ra, et al. Conformational changes in the M2 muscarinic receptor induced by membrane voltage and agonist binding. The Journal of physiology. 2011;589(Pt 7):1741–1753. doi: 10.1113/jphysiol.2010.204107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nature reviews. Drug discovery. 2006;5(12):993–6. doi: 10.1038/nrd2199. Available at: http://www.ncbi.nlm.nih.gov/pubmed/17139284. [DOI] [PubMed] [Google Scholar]
- Pardo L, et al. The role of internal water molecules in the structure and function of the rhodopsin family of G protein-coupled receptors. ChemBioChem. 2007;8(1):19–24. doi: 10.1002/cbic.200600429. [DOI] [PubMed] [Google Scholar]
- Pert CB, Synder SH. Opiate Receptor Binding of Agonists and Antagonists Affected Differentially by Sodium. Molecular pharmacology. 1974;10(6):868–879. [Google Scholar]
- Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nature reviews. Molecular cell biology. 2002;3(September):639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- Quitterer U, et al. Na+ ions binding to the bradykinin B2 receptor suppress agonist-independent receptor activation. Biochemistry. 1996;35(41):13368–77. doi: 10.1021/bi961163w. Available at: http://www.ncbi.nlm.nih.gov/pubmed/8873604. [DOI] [PubMed] [Google Scholar]
- Ranganathan A, Dror RO, Carlsson J. Insights into the Role of Asp792.50 in β2 Adrenergic Receptor Activation from Molecular Dynamics Simulations. Biochemistry. 2014;53(46):7283–7296. doi: 10.1021/bi5008723. Available at: http://pubs.acs.org/doi/abs/10.1021/bi5008723. [DOI] [PubMed] [Google Scholar]
- Rask-Andersen M, Masuram S, Schiöth HB. The Druggable Genome: Evaluation of Drug Targets in Clinical Trials Suggests Major Shifts in Molecular Class and Indication. Annual Review of Pharmacology and Toxicology. 2014;54(1):9–26. doi: 10.1146/annurev-pharmtox-011613-135943. Available at: http://www.ncbi.nlm.nih.gov/pubmed/24016212. [DOI] [PubMed] [Google Scholar]
- Rasmussen SGF, et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. [Accessed May 5, 2014];Nature. 2011 477(7366):549–55. doi: 10.1038/nature10361. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3184188&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinne A, et al. The mode of agonist binding to a G protein-coupled receptor switches the effect that voltage changes have on signaling. Science Signaling. 2015;8(401):ra110–ra110. doi: 10.1126/scisignal.aac7419. Available at: http://stke.sciencemag.org/cgi/doi/10.1126/scisignal.aac7419. [DOI] [PubMed] [Google Scholar]
- Šali A, Blundell TL. Comparative Protein Modelling by Satisfaction of Spatial Restraints. Journal of Molecular Biology. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. Available at: http://www.ncbi.nlm.nih.gov/pubmed/8254673%5Cnhttp://www.ncbi.nlm.nih.gov/pubmed/8254673?ordinalpos=1&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_DefaultReportPanel.Pubmed_RVDocSum. [DOI] [PubMed] [Google Scholar]
- Selent J, et al. Induced effects of sodium ions on dopaminergic G-protein coupled receptors. [Accessed April 24, 2014];PLoS Computational Biology. 2010 6(8) doi: 10.1371/journal.pcbi.1000884. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2920834&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selley DE, et al. Effects of sodium on agonist efficacy for G-protein activation in mu-opioid receptor-transfected CHO cells and rat thalamus. British journal of pharmacology. 2000;130(5):987–996. doi: 10.1038/sj.bjp.0703382. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10882382%5Cnhttp://www.ncbi.nlm.nih.gov/pmc/articles/PMC1572152/pdf/130-0703382a.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, et al. Mechanistic insights into the allosteric modulation of opioid receptors by sodium ions. [Accessed July 31, 2014];Biochemistry. 2014 53(31):5140–9. doi: 10.1021/bi5006915. Available at: http://www.ncbi.nlm.nih.gov/pubmed/25073009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen ARB, et al. GPCR-G Protein-B-Arrestin Super-Complex Mediates Sustained G Protein Signaling. Cell. 2016;166(4):907–919. doi: 10.1016/j.cell.2016.07.004. Available at: http://dx.doi.org/10.1016/j.cell.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanni S, et al. A Conserved Protonation-Induced Switch can Trigger “Ionic-Lock” Formation in Adrenergic Receptors. Journal of Molecular Biology. 2010;397(5):1339–1349. doi: 10.1016/j.jmb.2010.01.060. Available at: http://dx.doi.org/10.1016/j.jmb.2010.01.060. [DOI] [PubMed] [Google Scholar]
- Venkatakrishnan aJ, et al. Molecular signatures of G-protein-coupled receptors. [Accessed March 21, 2014];Nature. 2013 494(7436):185–94. doi: 10.1038/nature11896. Available at: http://www.ncbi.nlm.nih.gov/pubmed/23407534. [DOI] [PubMed] [Google Scholar]
- Vickery ON, Machtens J-P, Tamburrino G, et al. Structural Mechanisms of Voltage Sensing in G Protein-Coupled Receptors. Structure. 2016;24(6):997–1007. doi: 10.1016/j.str.2016.04.007. Available at: http://linkinghub.elsevier.com/retrieve/pii/S0969212616300454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickery ON, Machtens J-P, Zachariae U. Membrane potentials regulating GPCRs: insights from experiments and molecular dynamics simulations. Current Opinion in Pharmacology. 2016;30:44–50. doi: 10.1016/j.coph.2016.06.011. Available at: http://dx.doi.org/10.1016/j.coph.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf MG, et al. G-membed: Efficient insertion of a membrane protein into an equilibrated lipid bilayer with minimal perturbation. Journal of Computational Chemistry. 2010;31:2169–2174. doi: 10.1002/jcc.21507. [DOI] [PubMed] [Google Scholar]
- Yuan S, et al. Activation of G-protein-coupled receptors correlates with the formation of a continuous internal water pathway. Nature Communications. 2014;5(May):4733. doi: 10.1038/ncomms5733. Available at: http://www.nature.com/doifinder/10.1038/ncomms5733. [DOI] [PubMed] [Google Scholar]
- Yuan S, et al. Mechanistic Studies on the Stereoselectivity of the Serotonin 5-HT 1A Receptor. Angewandte Chemie International Edition. 2016;55(30):8661–8665. doi: 10.1002/anie.201603766. Available at: http://doi.wiley.com/10.1002/anie.201603766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, et al. High-resolution crystal structure of human protease-activated receptor 1. Nature. 2012;492(7429):387–92. doi: 10.1038/nature11701. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3531875&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Hummer G. Drying transition in the hydrophobic gate of the GLIC channel blocks ion conduction. Biophysical Journal. 2012;103(2):219–227. doi: 10.1016/j.bpj.2012.06.003. Available at: http://dx.doi.org/10.1016/j.bpj.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.