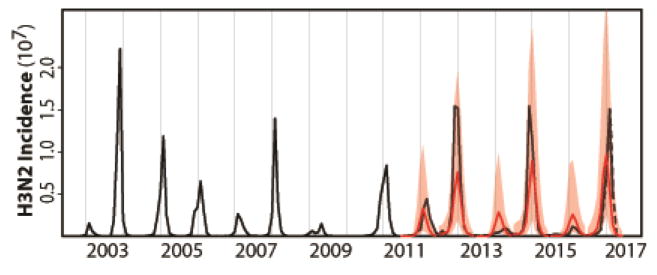
Figure 3.
H3N2 incidence forecasts based on the cluster model for the US. Both retrospective forecasts (for each influenza season from 2011/2012 to 2015/2016) and a real forecast for the coming 2016/2017 influenza season are represented. These forecasts are simulated on a seasonal basis from estimated initial conditions starting in June and based on parameters estimated with all the data up to that point in time. The average monthly H1N1 incidence from this training dataset was used for forecasting purposes as the observation of this driver quantity would not be available. Similarly, the quantities specifying the evolutionary change of the virus was extrapolated as the sequences required for their computation would not be available. The black curve is the monthly observed H3N2 incidence; the red curve is the predicted monthly median incidence with shaded 2.5–97.5% quantiles from 1000 random simulations with the best models. The cluster model captures the occurrence of low and high seasons and forecasts high H3N2 incidence risk level for the 2016/2017 influenza season. The observed incidence data for the 2016/2017 influenza season, which were not yet available when this study was conducted, are shown with the dotted line (and based on data from the weekly US influenza surveillance report until week 14 ending on April 8, 2017).