Abstract
A limited number of studies have been conducted to analyze ribosomal RNA (rRNA, present in the ribosome) in bioaerosol samples to identify currently or potentially active airborne microbes, although its genomic counterpart, the rRNA gene (on the chromosome) has been frequently targeted for airborne microbial community analysis. A knowledge gap still exists regarding whether the bioaerosol rRNA abundances are affected by the bioaerosol collection process. We investigated the effect of air sampling stress on the measurement and characterization of 16S rRNA for bioaerosols in the laboratory and field experiments using quantitative polymerase chain reaction (qPCR) and high-throughput sequencing techniques. In a laboratory study, known quantities of freshly grown Escherichia coli cells were spiked onto the filter of a Button Aerosol Sampler and into liquids of BioSampler and SpinCon air samplers and then exposed to sampling stress when the samplers were operated for 2 hours. We found that the recovered cellular 16S rRNA abundance as determined by qPCR was dependent on sampler type. Further, two devices (Button Aerosol Sampler and BioSampler) that exhibited markedly different efficiency in preserving 16S rRNA were employed in an outdoor environment to collect bioaerosols simultaneously on eight days in two different seasons. The abundance of 16S rRNA in the outdoor air sample (1.3×106–4.9×107 copies/m3) was about two orders of magnitude higher than that of 16S rRNA gene (6.9×103–1.5×105 copies/m3). The 16S rRNA sequences revealed a different bacterial community compared with 16S rRNA gene-based results across all samples, and this difference depended on the sampling device. In addition, a number of bacterial taxa exhibited higher abundance in the 16S rRNA gene sequences than in 16S rRNA sequences, which suggests the potential activities of certain microbes in airborne phase. Overall, this study highlights the importance of sampling device selection when analyzing RNA in bioaerosols.
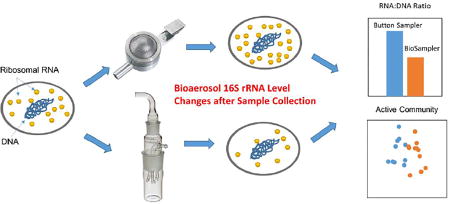
Graphical abstract
INTRODUCTION
The ribosomal RNA (rRNA) gene (on the microbial chromosome), particularly the 16S rRNA gene of Bacteria and/or Archaea, has been widely utilized in environmental microbiology studies because it is ubiquitous and has low mutation rates throughout Bacteria and Archaea evolution1, and it also encompasses hypervariable regions which can be used to distinguish between bacterial taxa2. However, microbial samples often contain 16S rRNA genes from dormant cells3, 4 or lysed cells5, 6, thus precluding the use of 16S rRNA gene when investigating only active microbial community members. In contrast, the rRNA (as part of the ribosome) encoded by this gene is directly linked to cell physiology, e.g. the synthesis of rRNA is growthrate dependent for a number of bacterial species7–9. Thus, analysis of 16S rRNA sequences rather than rRNA gene sequences (DNA) can help reveal those community members who are or have recently been active within a complex microbial community10, 11. This approach has been employed to analyze microbial samples from water, soil, sediments, and biofilms10–17. However, there has been a limited number of studies on rRNA measurements and rRNA-based community analysis in airborne microorganisms (a.k.a. bioaerosols)18, 19.
Since airborne biomass is relatively low in abundance compared to soil or water biomass20, 21, bioaerosol sampling devices often have to be operated for long time periods which could cause stress on the already collected microorganisms as sample collection continues22–24. Thus, one major concern when studying rRNA in bioaerosols is whether the rRNA abundance in the collected cells remains unchanged during sampling. Although rRNA is a relatively stable RNA type, it could also exhibit variation within a cell under certain changes in environmental conditions25. It has been reported that bacterial rRNA concentrations increase during early exponential growth of cells7 and rRNA degrades when cells experience depletion of nutrients and glucose starvation25. A recent study showed that the 16S rRNA abundance in Sphingomonas aerolata aerosols in a rotating bioreactor increased when the bacteria were supplied with gaseous growth substrates18. Certain bacteria, e.g. Lactococcus lactis in a non-growth state displayed changes in rRNA content in response to heat shock26. Since air sampling processes such as impaction, impingement, and desiccation can significantly alter the physiological status of collected bioaerosols, including the loss of viability and impaired cell membrane integrity22–24, 27, it is possible that air sampling also affects the abundance of rRNA in the samples and introduces bias to rRNA-based sample analysis. Thus, if rRNA is to be used to analyze airborne microbial communities, it is important to determine how the sampling process, i.e., stress due to sampling, affects the rRNA of bioaerosol samples and whether the magnitude of the effect depends on a particular bioaerosol collection method or device.
Historically, the growth or metabolic activity of particular bacterial taxa has been investigated by measuring the change in 16S rRNA:16S rRNA gene ratio7–9, 28. Subsequently, 16S rRNA sequence analysis has been used to identify potentially active members within microbial populations from complex microbial samples11–16. However, if the sampling process itself, e.g. sampling stress, leads to the increase or decrease in rRNA content of specific bacterial taxa, then their relative abundance within a complex bacterial community could be either overestimated or underestimated. Because the different particle capturing mechanisms of various sampler designs may cause differing effects on cellular rRNA, this potential effect on the sequence abundance of active microbial community members may be air sampling device-dependent.
The objectives of this study were: 1) to study whether the rRNA content of bioaerosol samples changes due to the air sampling process itself, i.e., sampling stress; 2) to assess whether this effect of sampling stress on bioaerosol rRNA is device-dependent; 3) to investigate how this effect impacts the analysis of 16S rRNA sequences from bioaerosols collected in an outdoor environment. First, we investigated the change in 16S rRNA:16S rRNA gene ratio of Escherichia coli in response to two-hour aerosol sampling using three different bioaerosol samplers in a laboratory. In the second part of the study, we analyzed microbial communities simultaneously collected from the outdoor air by the same three devices on eight different days in summer and late winter/early spring. The microbial communities represented in the 16S rRNA gene and 16S rRNA were analyzed by pyrosequencing. To the best of our knowledge, this is the first study to investigate the potential effect (bias) of sampling stress on the quantification and characterization of 16S rRNA from bioaerosol samples.
MATERIALS AND METHODS
Bacterial Culture in Laboratory Experiments
A Gram-negative bacterium E. coli (ATCC 15597, Manassas, VA) was selected as a test microorganism. This organism has been used as a model microorganism in many bioaerosol studies23, 27, 29. The procedures for preparation of E. coli suspension were described elsewhere30 and details are provided in Supplementary Information. Briefly, E. coli was precultured in Tryptic Soy broth, harvested by centrifugation, and resuspended in 1×phosphate-buffered saline (PBS) solution prior to experiments.
Bioaerosol Samplers
A Button Aerosol Sampler (SKC Inc., Eighty Four, PA), later referred to as Button sampler, a BioSampler (SKC Inc.), and a SpinCon wet cyclone (PAS 450-10A, InnovaPrep LLC., Drexel, MO) were used in this study to collect bioaerosols. The Button sampler is a filter-based sampler, and it was used with a 0.8-µm-pore-size polyethersulfone (PES) membrane filter (SUPOR, Pall Life Sciences, Port Washington, NY). Its nominal flow rate is 4 liters/min31, but here the sampler was operated at a flow rate of 18 liters/min to exacerbate a potential effect of filtration stress on bacterial cells. The two other devices are liquid-based bioaerosol samplers and collect airborne particles by a combination of liquid impingement and cyclonic action. 1×PBS solution was used as collection fluid for these two devices. The SKC BioSampler with a 5-ml collection cup was operated at its design flow rate of 12.5 liters/min. The BioSampler cup was refilled with Milli-Q water approximately every 15 minutes to replenish the fluid evaporated during its operation. The SpinCon air sampler was operated at a flow rate of 450 L/min and the total sample volume of approximately 10 ml; the device automatically maintains liquid level during its operation from a reservoir of sterile water.
Experimental Procedure
In order to simulate an environment where bacterial cells were continuously exposed to sampling stresses after their initial collection, each device was spiked with a known amount of bacteria and then operated for 2 hours at a room temperature by aspirating particle-free air inside a disinfected class II biosafety cabinet (NuAire Inc., Plymouth, MN). The air temperature and relative humidity (RH) for tested conditions were 25 °C and between 25–30%, respectively. The following amounts of freshly grown E. coli cells in 1×PBS were spiked prior to sampling: ~5×105 cells were loaded onto the filter, ~3×108 cells were spiked into the collection fluid inside the BioSampler cup, and ~1×109 cells were added to the SpinCon collection chamber. A separate aliquot of the same E. coli cells was saved at −80°C to serve as a reference sample.
After the samplers had been operated for 2 hours, the filter was removed from the Button sampler with sterile tweezers, immediately placed into a sterile 1.5 ml microcentrifuge tube and stored at −80°C. The whole filter was subjected to subsequent nucleic acid extraction within the microcentrifuge tube. One ml of homogenized liquid was taken from the BioSampler and SpinCon samples and 10 µl β-mercaptoethanol (β-ME) was added to inhibit the potential RNase activity. The liquid samples were centrifuged at 16,100×g for 10 min at 4°C, after which 950 µl of the supernatant liquid was transferred into a new 1.5 ml centrifuge tube and saved along with the rest 50 µl of the sample containing pellet cells at −80°C. Our previous study showed that the supernatant liquid after centrifugation of liquid-based bioaerosol samples could have a substantial quantity of extracellular DNA from membrane damaged cells23. Thus, both fractions of samples were subjected to subsequent DNA and RNA analysis.
The outdoor sampling experiments were performed with Button Aerosol Sampler and BioSampler on Rutgers University’s Cook Campus in New Brunswick, NJ (40.48°N, 74.44°W). Here, we chose only two types of devices for investigation because they exhibited markedly different efficiencies in preserving 16S rRNA from initial laboratory investigation with E. coli, while the SpinCon did not show significant impact on the recovery of E. coli 16S rRNA (explained with details in Results section). The sampling location was on a grass field and about 10 meters from a teaching and research building. One Button sampler and two BioSamplers were located approximately 1 m above the ground and each sampler set approximately 0.5 m from any other sampling devices, i.e. the samplers were collocated. The Button samplers have omnidirectional inlet and are only minimally affected by wind direction. All three sampling devices were operated simultaneously for two hours on three different days in summer (Aug. 6th, Sep. 15th, and 17th of 2014) and five different days in late winter/early spring (Feb. 4th, Mar. 13th, 16th, 23rd, and 25th of 2015). Two BioSampler units were employed here to collect sufficient biomass for subsequent analysis. The temperature during sampling varied between 21–26°C in summer and 3–12°C in late winter/early spring. The RH during sampling was between 60–70% in summer and 23–55% in late winter/early spring.
Upon completion of each sampling event, the filter from the Button sampler was removed with sterile tweezers, immediately placed into a 1.5 ml sterile microcentrifuge tube and stored at −80°C for subsequent nucleic acid extraction. The collection liquid from the two BioSamplers was combined and transferred into a 50-ml centrifuge tube. Then two BioSamplers were refilled with 2 ml sterile water each and shaken vigorously to wash the residual particles off inner surfaces32. The washed liquid was then combined with the initial sample reaching a total volume of ~15 ml. A 1% β-ME solution was added. Thereafter, the liquid sample was centrifuged at 16,100×g for 10 min at 4°C. After centrifugation, the pellets were immediately saved at −80°C. The supernatant liquid was first extracted with sec-butanol (Acros Organics, Somerset, NJ) to reduce the sample volume to ~400 µl and then stored at −80°C. A blank filter for Button sampler and sterile liquid solutions for BioSampler were saved and analyzed for quality control.
Nucleic Acids Extraction
The total genomic DNA and RNA of E. coli laboratory samples and outdoor bioaerosol samples from cell pellets and supernatant lipid were extracted by a phenol-chloroform method as described elsewhere18. For analysis of 16S rRNA, the extracted nucleic acids mixtures were subjected to DNase treatment followed by reverse transcription. The details of nucleic acids extraction and 16S rRNA preparation protocols are provided in Supplementary Information.
Quantitative PCR
We previously developed a dual-internal-reference technique to improve the accuracy of analysis when quantifying bacterial 16S rRNA:16S rRNA gene ratio by introducing two exogenous DNA and RNA references (Pseudomonas fluorescens genomic DNA and 16S rRNA)30. This technique was applied here to quantify the 16S rRNA and 16S rRNA gene for E. coli samples from the laboratory experiment by using a multiplex qPCR method30. The 16S rRNA and 16S rRNA gene from bioaerosols collected outdoors were quantified by using a SYBR-Green-based qPCR assay that was performed on iCycler iQ5 RT-PCR detection system (Bio-Rad Laboratories, Hercules, CA) by following a previously reported method23. The details of both qPCR assays are provided in Supplementary Information.
Sequence Analysis
Microbial communities in outdoor samples were characterized by a commercial laboratory (Molecular Research LP, Shallowater, TX) using multiplex barcoded 16S rRNA amplicon pyrosequencing. Prior to sequencing, the bacterial 16S rRNA gene sequences and reversed transcribed 16S rRNA sequences in each sample were amplified by PCR with universal primer sets 515f/909r. All amplicon products from different samples were mixed in equal concentrations and purified using Agencourt Ampure beads (Agencourt Bioscience Corporation, MA, USA). The pyrosequencing was performed on a Roche 454 FLX+ titanium instrument (454 Life Sciences, Branford, CT) following manufacturer’s guidelines and reagents.
The obtained sequences were analyzed using the Quantitative Insights Into Microbial Ecology (QIIME) software package33. Quality filtering was performed by removing any sequences with less than 200 base pairs in length, machine quality score lower than 25, containing any mismatches in the barcode or primer sequence, or having any ambiguous bases. Chimeric sequences were removed by UCHIME34. A total of 1.8×105 sequences passed the quality control checks and were subjected to subsequent analyses. Sequences were clustered into operational taxonomic units (OTUs) by UCLUST35 using minimal 97% sequence similarity, and the representative sequence of each OUT was aligned with PyNAST33 against the Greengenes core set from July 201236. Taxonomic assignment was conducted using the Ribosomal Database Project classifier37. All samples were rarefied to 702 sequences prior to downstream analyses of diversity and community composition to correct for different sequencing depth. The relative abundance of a bacterial taxon was defined as the number of sequences affiliated to the particular taxon divided by the total number of sequences per sample. The phylogenetic distance in the microbial community between paired samples was analyzed using weighted UniFrac algorithm38, and the results were presented in principal coordinate analyses (PCoA) plots. Sequences were deposited in the NCBI Sequence Read Archive database under accession number SRP112875.
Statistical Analysis
For each device in laboratory experiments, a one-way ANOVA with Fisher’s LSD analysis was performed to compare 16S rRNA gene and 16S rRNA quantities, and 16S rRNA:16S rRNA gene ratios for E. coli samples before and after two hours of sampling. For outdoor experiments, a two-way nested ANOVA was performed to test the effects of sampling device and season on 16S rRNA gene and 16S rRNA quantities, and their ratio. A random factor, sampling day, was nested under the main factor season in the two-way ANOVA model to stratify the data. Three-way nested ANOVA test was performed to test the effects of season (nesting sampling days), sampling device and sequence types on the number of identified bacterial genera, and the sum of abundances of bacterial genera detected in both 16S rRNA and 16S rRNA gene sequences in each sample. Permutational ANOVA (PERMANOVA) was performed in R (ADONIS function in VEGAN package, version 2.4–3)39 to test the effects of season, sequence type (16S rRNA or rRNA gene), sampling device and the interactions between sequence type and sampling device on the weighted-UniFrac pairwise distances of bacterial communities. We performed a Mann-Whitney U test using SPSS (version 20, IBM Corp., Armonk, NY) to test the seasonal effect on the relative abundance of individual detected bacterial families and genera and the ratios of 16S rRNA: 16S rRNA gene across all bacterial families. A Wilcoxon Signed Rank test was performed using SPSS to compare the relative abundance of individual detected bacterial family/genus between paired 16S rRNA gene and 16S rRNA sequences. Overall, a statistically significant difference was assumed for p<0.05.
RESULTS
Effect of Sampling Device on 16S rRNA:16S rRNA Gene Ratio of E. coli
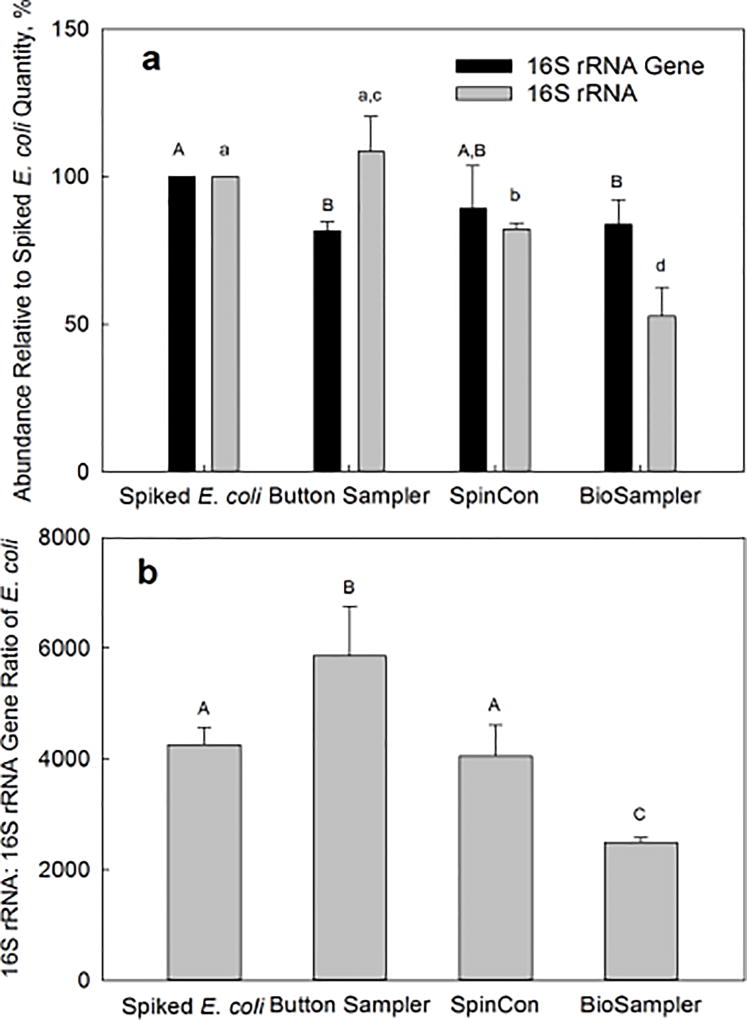
Figure 1a shows a change in the quantity of spiked E. coli 16S rRNA and 16S rRNA gene after two hours of sampling in laboratory experiments as a function of the sampling device. It should be noted here that the 16S rRNA and 16S RNA gene abundances in E. coli cells were determined with qPCR by targeting the same section of the E. coli 16S RNA gene sequence, and the results were reported as percentage recovery relative to the quantities of 16S rRNA or 16S rRNA gene in spiked cells, respectively. The recovered 16S rRNA gene quantities were 81.7±3.1%, 89.2±14.6 % and 84.0±8.2% for Button sampler, SpinCon sampler, and BioSampler, respectively. This suggests a relatively low and similar loss of initially collected E. coli cells during continuing sample collection for two hours. However, the differences are more marked for the recovery of 16S rRNA: 108.5±12.0%, 82.3±2.1% and 52.8±9.7% for Button sampler, SpinCon air sampler, and BioSampler, respectively. The distinct difference in the relative abundance of 16S rRNA among the devices is also reflected in the 16S rRNA:16S rRNA gene ratios (Figure 1b). For the Button sampler, the ratio increased significantly (p=0.034) from ~4300 to ~5900 after two hours of particle-free air sampling. For E. coli spiked into liquid-based samplers, the ratio associated with SpinCon sampler remained steady at ~4000 (p=0.44); however, two hours of active aspiration with particle-free air by the BioSampler resulted in an approximately 40% decrease (p=0.026) in the 16S rRNA:16S rRNA gene ratio relative to that of the initially spiked E. coli cells: from 4330 to 2500.
Figure 1.
The effect of sampling device on the 16S rRNA:16S rRNA gene ratio for E. coli cells spiked into the three samplers (Button sampler, SpinCon sampler, and BioSampler) and recovered after two hours of active sampling of particle-free air. Each bar group (or bar) from left to right shows the average for 10, 4, 3 and 3 samples respectively; error bars are one standard deviation. Bars with different capital letters (A or B) or small letters (a, b, c or d) are statistically different (Fisher’s LSD, p<0.05). a) The abundance of 16S rRNA gene and 16S rRNA recovered from each sampler compared to an initially spiked reference quantity of 16S rRNA gene or 16S rRNA. b) The 16S rRNA:16S rRNA gene ratio of E. coli cells spiked into three samplers and recovered after two hours of active air sampling of particle-free air compared with the ratio of initially spiked E. coli cells.
Quantification of Outdoor Bioaerosols
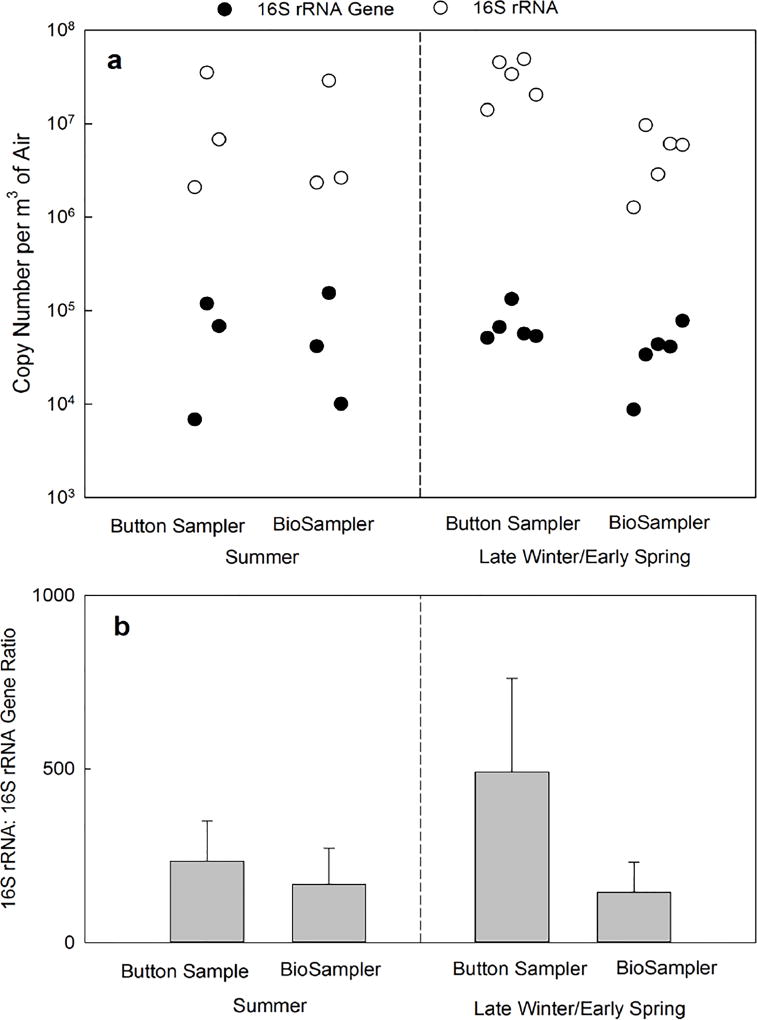
Similar to the laboratory experiment, the 16S rRNA and 16S RNA gene abundances in bioaerosols collected outdoors were determined using qPCR by targeting the same section of bacterial 16s RNA gene sequence, and the results were reported as absolute copy number of PCR amplicons. The abundance of 16S rRNA gene ranged from 6.9×103 to 1.2×105 copies/m3 and from 1.0×104 to 1.5×105 copies/m3 for outdoor bioaerosols collected in summer by Button sampler and BioSampler, respectively (Figure 2a). In late winter/early spring samples, the abundance of 16S rRNA gene ranged from 5.1×104 to 1.3×105 copies/m3 for the Button sampler and from 8.8×103 to 7.8×104 copies/m3 for the BioSampler. No significant differences were observed between 16S rRNA gene quantities in samples collected by the two samplers or in two different seasons.
Figure 2.
a) The abundance of bacterial 16S rRNA gene (closed circle) and 16S rRNA (open circle) in air samples collected by Button sampler and BioSampler simultaneously in an outdoor environment for two hours on three different days in summer (left) and five different days in late winter/early spring (right). The paired 16S rRNA gene and 16S rRNA data points are aligned vertically and positioned on the x-axis in order of sampling date. b) The 16S rRNA:16S rRNA gene ratio of bioaerosol samples collected by Button sampler and BioSampler simultaneously in an outdoor environment for two hours on three different days in summer (left) and five different days in late winter/early spring (right). The bars are averages and error bars show one standard deviation.
In general, the 16S rRNA abundance was about two orders of magnitude higher than that of 16S rRNA gene copies from the same sample. For example, the summer samples collected by Button sampler and BioSampler contained a range of 2.1×106–3.5×107 copies/m3 and 2.3×106–2.9×107 copies/m3 of 16S rRNA (Figure 2a). For late winter/early spring samples, the abundance of 16S rRNA ranged from 1.4×107 to 4.9×107 copies/m3 for the Button sampler and from 1.3×106 to 9.6×106 copies/m3 for the BioSampler. ANOVA test indicated that the 16S rRNA quantities collected by BioSampler were significantly lower than those collected by Button sampler (p=0.001).
The 16S rRNA:16S rRNA gene ratios of collected bioaerosols were 234±116 and 168±104 for Button sampler and BioSampler from three summer sampling events, and 491±270 and 144±87 for Button sampler and BioSampler in five late winter/early spring samples, respectively (Figure 2b). The 16S rRNA:16S rRNA gene ratios from BioSampler samples were found to be significantly lower than those from Button sampler (p=0.04) by ANOVA.
Comparison in Airborne Microbial Community
Taxonomic identification of paired 16S rRNA gene and 16S rRNA sequences for all sixteen outdoor samples was completed at the genus level or higher due to the short reading lengths (466 bp) obtained in pyrosequencing. The Proteobacteria was the most abundant bacterial phylum on average, and it accounted for 21.7%, 5.8% and 10.2% of all reads for α-, β- and γ- subgroups (Figure S1 in Supplementary Information), respectively. Other dominant phyla included the Actinobacteria (17.6%), Bacteroidetes (11.6%), Cyanobacteria (9.1%) and Firmicutes (9.8%). The relative abundance of each bacterial phylum and class identified in all outdoor samples are presented in detail in Table S1 (Supplementary Information).
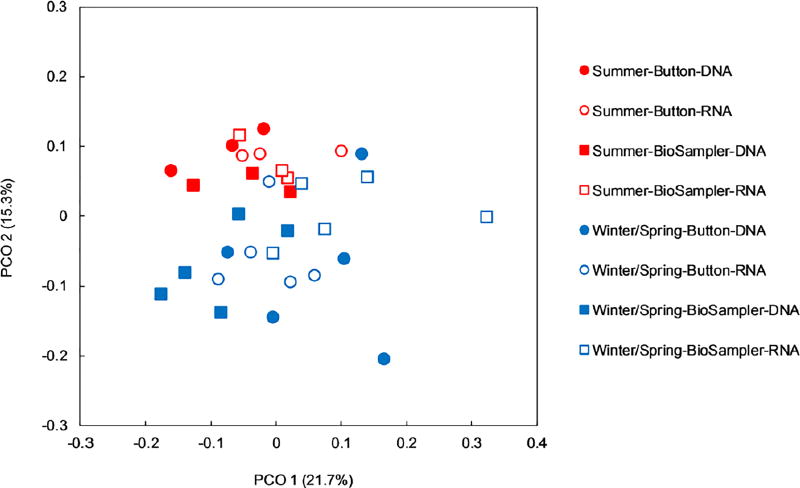
A principle coordinate analysis (PCoA) of weighted-UniFrac distance was conducted to compare the bacterial communities across all outdoor samples, and the results are presented in Figure 3. The data points representing summer samples were clustered together and clearly separated from the data points for late winter/early spring. There was no distinct clustering of sample points separated by either sampling device (Button sampler or BioSampler) or sequence type (16S rRNA gene sequences or 16S rRNA sequences). Results from permutational ANOVA (PERMANOVA) analysis (Table 1) showed that bacterial communities were significantly different between two seasons (p=0.011) and also between two sequence types (p=0.006), while no difference was observed in bacterial communities collected by the two devices (p=0.869). Interestingly, the interaction between sampling device and sequence type exhibited significant impact (p=0.046) on the bacterial community composition.
Figure 3.
Weighted UniFrac-based bacterial diversity principal coordinate analysis of outdoor air samples collected by two devices (Button sampler: round markers; BioSampler: square markers) in two seasons (summer: red markers; late winter/early spring: blue markers) and analyzed based on 16S rRNA gene (DNA, filled markers) and 16S rRNA sequences (RNA, open markers).
Table 1.
PERMANOVA results on the effects of season, sampling device, sequence type, as well as the interaction between sampling device and sequence type on the bacterial community weighted-UniFrac pairwise distances. Bold text indicates that p< 0.05.
| Variables | df | Pseudo-F | p |
|---|---|---|---|
| Season | 1 | 3.841 | 0.011 |
| Sampling Device | 1 | 0.551 | 0.869 |
| Sequence Type | 1 | 2.157 | 0.006 |
| Sampling Device × Sequence Type | 1 | 1.582 | 0.046 |
| Residuals | 27 | 0.769 | |
| Total | 31 |
The relative abundance of major bacterial families (>1% average abundance in samples from either season) was further compared between two seasons, and those bacterial families that exhibited significantly different abundances are shown in Figure S2 (Supplementary Information). The Sphingomonadaceae, Methylobacteriaceae, Sphingobacteriaceae, Enterobacteriaceae, and Pseudonocardiaceae families were more abundant in summer samples relative to late winter/early spring samples. The genera Methylobacterium (Methylobacteriacea family), Novosphingobium (Sphingobacteriaceae family) and Actinomycetospora (Pseudonocardiaceae family) showed the greatest differences between two seasons: their average abundances in summer compared to late winter/early spring samples were higher by factors of 4, 4, and 10 respectively. On the contrary, families Moraxellaceae and Flavobacteriaceae showed higher abundance in late winter/early spring than in summer. The representative genera contributing to the difference are Psychrobacter (Moraxellaceae family) and Flavobacterium (Flavobacteriaceae family). The abundance of these two genera in late winter/early spring was higher than in summer by a factor of approximately 150x and 5x, respectively.
Comparison Between Relative Abundance of 16S rRNA and 16S rRNA Gene Sequences in Outdoor Bioaerosols
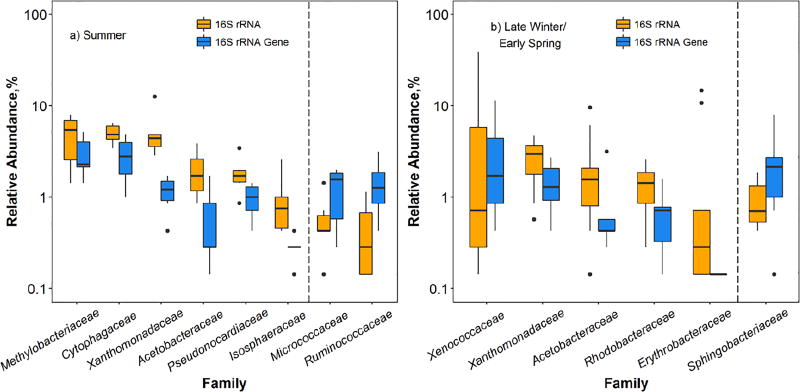
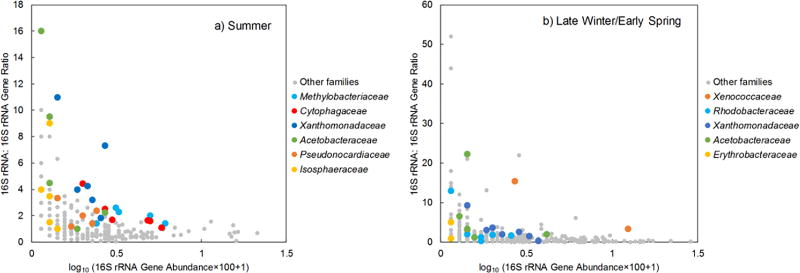
The 16S rRNA and 16S rRNA gene-based community data were further analyzed for differences at both family and genus levels. Because different seasons harbored distinctly different bacterial communities, the comparison was stratified by season. For summer samples (Figure 4a), bacterial families Methylobacteriaceae, Cytophagaceae, Xanthomonadaceae, Acetobacteraceae, Pseudonocardiaceae, and Isosphaeraceae showed a higher relative abundance of 16S rRNA than 16S rRNA gene. Within those families, genera that contributed to relatively higher 16S rRNA abundances were Methylobacterium (Methylobacteriaceae family), Hymenobacter (Cytophagaceae family), Rhodanobacter and Luteimonas (Xanthomonadaceae family), Roseomonas (Acetobacteraceae family), and Actinomycetospora (Pseudonocardiaceae family). For late winter/early spring samples (Figure 4b), families of Xenococcaceae, Xanthomonadaceae, Acetobacteraceae, Rhodobacteraceae, and Erythrobacteraceae exhibited a significantly higher relative abundance of 16S rRNA than that of 16S rRNA gene. At the genus level, genera Rhodanobacter (Xenococcaceae family), Luteimonas (Xanthomonadaceae family), and Roseomonas (Acetobacteraceae family) were determined to have relatively higher abundances of 16S rRNA. On the other hand, two bacterial families (Micrococcaceae and Ruminococcaceae) in summer samples and one family (Sphingobacteriaceae) in late winter/early spring samples exhibited a higher relative abundance of 16S rRNA gene than 16S rRNA (Figure 4). No particular bacterial genus was detected in greater abundance in 16S rRNA gene than 16S rRNA in samples collected from either season.
Figure 4.
Bacterial genera that exhibited different relative abundances (p<0.05) in 16S rRNA and 16S rRNA gene sequences of outdoor bioaerosols collected in two seasons. Each boxplot represents the results of six samples collected in a) summer and ten samples collected in b) late winter/early spring. To the left of the dashed line: bacteria genera showing higher relative abundance in 16S rRNA sequences than in 16S rRNA gene sequences; to the right of the dashed line: bacteria genera exhibiting lower relative abundance in 16S rRNA sequences than in 16S rRNA gene sequences. In each boxplot, the line through the middle represents the median. The bottom and top of each box are the 25th and 75th percentiles. Outliers are indicated with points beyond the whiskers and are defined as the data points lower (or higher) than 1.5 interquartile (the difference between the upper and lower quartiles) range of the lower (or higher) quartile.
A positive correlation was observed between the log-transformed relative abundances of 16S rRNA and 16S rRNA gene across all bacterial genera. (Figure S3 in Supplementary Information, Pearson correlation p<0.0001, R2=0.22). The 16S rRNA:16S rRNA gene ratios were calculated with the relative abundances of bacterial families that were found in both the paired 16S rRNA and 16S rRNA gene sequences, and the ratios ranged from 0.07 to 16 (1.44 on average) and from 0.02 to 52 (2.00 on average) for samples collected in summer and late winter/early spring, respectively. No difference was found between the ratios of all bacterial families from two seasons (p=0.13). Negative correlations (Figure 5) were observed between the 16S rRNA:16S rRNA gene ratio and relative abundances of 16S rRNA gene of bacterial families across all samples from two seasons (Kendall correlation p<4e–11, τ= −0.27 for summer, p<2e–16, τ= −0.35 for late winter/early spring samples).
Figure 5.
The relationship between 16S rRNA:16S rRNA gene ratio and relative abundance of 16S rRNA gene in a) summer and b) late winter/early spring air samples collected in an outdoor environment. Each point represents individual bacterial family detected in one sample. Colored points represent those bacterial families that exhibited significantly higher relative abundances of 16S rRNA than 16S rRNA gene.
DISCUSSION
It has been known that microbes in an active growth stage may shift towards the non-growth maintenance activities under certain stress11, 40. For example, E. coli was reported to actively respond to desiccation stress by changing the membrane phase behavior41–43, e.g. increasing the fraction of saturated fatty acids41, and synthesizing more intracellular compatible organic solutes including trehalose, proline and glutamine42, 43. At the same time, only a limited amount of published work is currently available regarding the relationship between non-growth-related cell activities and rRNA concentration11, 26. In our tests, E. coli cells exposed to osmotic and desiccation stress did not manifest any growth: a lower quantity of genomic DNA was recovered from filters post-exposure relative to the initial quantity in spiked E. coli cells. Thus, non-growth cell maintenance of E. coli may partially explain the elevated 16S rRNA:16S rRNA gene ratio under desiccation conditions.
In our tests with two liquid-based samplers, a significant degradation of 16S rRNA was observed in E. coli samples after two hours of particle-free air sampling with the BioSampler, but not with the SpinCon sampler, although both devices use a similar collection principle. It is worth mentioning that we observed a greater temperature drop in the collection liquid of BioSampler after 2 hours of operation (25 °C to 12 °C) than in SpinCon (25 °C to 22 °C). Cold shock to E. coli has been known to elevate the RNase R activity within E. coli cells44, 45, e.g. more than 10-fold increase was observed for a temperature drop from 37°C to 10°C46. Thus, we hypothesize that this difference in the temperature drop might be the main factor responsible for different rRNA abundance in samples from the two devices. For both devices, the air is pulled through either three nozzles (BioSampler) or a thin slot (SpinCon) at high velocities and then impinged into the liquid inside the collection vessel. An air pressure below atmospheric is needed inside both collectors to create the air flow. Low air pressure results in a rapid evaporation of water, and thus the evaporative cooling effect on the remaining collection liquid47. However, each BioSampler nozzle operates as a critical orifice resulting in the air flow moving at the sonic velocity (~340 m/s)48 and pressure of about 0.5 atmospheres inside the collector; on the other hand, the air velocity through the SpinCon’s inlet slot is much lower at an estimated ~30 m/s (from manufacturer’s specifications), which results in a higher absolute pressure inside the SpinCon compared to BioSampler. Overall, the difference in absolute pressure inside the two samplers very likely leads to the faster liquid evaporation rate of BioSampler and thus a stronger cooling effect on the collection liquid containing bacteria than SpinCon.
In the laboratory experiments, statistically significantly higher fraction of spiked E. coli 16S rRNA was recovered from filter samples (Button sampler) than from BioSampler samples (Fig. 1). The same pattern held for total 16S rRNA recovered from outdoor samples (Fig. 2). Therefore, the potential impact of the two sampling devices on airborne bacterial community analysis was further investigated. While the outdoor community composition identified by 16S rRNA and 16S rRNA genes were statistically different from each other, there was no difference in community structure between samples collected by the two devices (Table 1). Interestingly, a significant interaction between the sampling device and sequence type was observed (p=0.046, Table 1), which suggests that the different community structure as revealed by 16S rRNA sequences compared to 16S rRNA gene sequences was dependent on sampling device. This finding, together with the result showing the sampler-dependent difference in 16S rRNA abundance, further highlights the importance of sampler selection when studying airborne bacteria using 16S rRNA sequences. However, unlike in our laboratory studies with E. coli, there was no reference sample that would have represented 16S rRNA sequences in outdoor bioaerosols prior to their collection. Thus, it was impossible to determine accurately how the air sampling stress changes the abundance of 16S rRNA of individual bacterial species and, consequently, the overall composition of the microbial community in outdoor air samples.
Our data show that bacterial communities collected at the same outdoor location were distinctly different in different seasons regardless of the sampling device and analysis sequence type (Figure 3 and Table 1). Other studies also reported temporal variation in airborne bacterial population across several consecutive days49 as well as across different seasons50–52. This temporal variability could be driven by changing contribution from individual sources, e.g. soils, water bodies, plant surfaces, animal and human activities51–53. For example, genera Methylobacterium54, 55, Novosphingobium56, 57 and Actinomycetospora58, 59 that are frequently associated with plant leaves were detected with higher abundances in summer samples than in late winter/early spring samples. In addition, meteorological conditions (temperature) also play a role in shaping the outdoor airborne microbial community60. For example, bacterial genera Psychrobacter and Flavobacterium were found with higher abundances in late winter/early spring samples than in those collected during summer. Both genera contain a number of psychrotolerant species and have been detected with higher frequency in low temperature and snow-covered environments50, 61–63.
In addition to the temporal variation in airborne bacterial population49, the selection of sampling device might also contribute to the observed microbial community difference across multiple samples. For instance, Hoisington et al.64 reported that four different types of active samplers that were collocated in a concurrent air sampling event revealed different microbial composition in an indoor environment. It was suggested that the unique design and working principle of various samplers might selectively capture microbes with different characteristics (e.g. aerodynamic diameter)64. Apart from this potentially influencing factor, it is also likely that the collocated samplers that operated simultaneously could introduce competing sampling artifacts due to the interference of air flow from other devices. In our experimental design, we deliberately adjusted the position of each sampler in order to minimize this potential artifact. In fact, we did not observe the variation in community composition between sampling devices (Table 1). However, it is possible that other main effects, e.g. temporal variation and sequence type (Table 1), might have masked the impacts from the two above-mentioned factors associated with sampler selection (the sampler type and competing air flow when placing side by side). As a result, the sampler-associated artifacts in bioaerosol characterization results should be considered and warrant future investigation.
Several bacterial families and genera were identified in the current study with higher relative abundance in 16S rRNA sequences than in 16S rRNA gene sequences (Figure 4). Among these bacterial taxa, the genera Hymenobacter (Cytophagaceae family) and Roseomonas (Acetobacteraceae family) were also recently reported with significantly higher 16S rRNA:16S rRNA gene ratios than other taxonomic groups in air samples collected at a high elevation research station18, 19. These findings suggest the prevalence of both bacterial genera in the atmosphere in a potentially active state across different geographic locations. However, considering the fact that the copy number of 16S rRNA genes in a bacterial genome varies among different species, the higher 16S rRNA:16S rRNA gene ratio does not necessarily indicate that the 16S rRNA content of a specific taxonomic group is elevated relative to that of others on a per-cell basis. For a specific bacterial species, the quantity of 16S rRNA per cell could be determined by multiplying the 16S rRNA:16S rRNA gene ratio by the copy number of 16S rRNA gene per genome. Therefore, for all bacterial genera that exhibited higher relative abundance in 16S rRNA than 16S rRNA gene in either summer or late winter/early spring samples, we searched the GenBank database (http://www.ncbi.nlm.nih.gov/genbank/) for their complete genomes. As a result, we found eleven, six and one complete genomes for Methylobacterium, Hymenobacter, and Rhodanobacter, respectively. No complete genome assembly could be retrieved for other bacterial genera. The Methylobacterium was the only genus that possessed a higher number of 16S rRNA gene copies (5.8 on average, Table S2 in Supplementary Information) in its genome than the average four copies per bacterial genome65, suggesting a greater current or potential cellular activity10, 11 of Methylobacterium spp. compared to other bacterial genera in summer. Bacteria from the genus Methylobacterium can grow on one-carbon compounds as the sole source of carbon and energy, and they have been reported in a variety of habitats including soil, dust, leaf surfaces and air, etc.55 As common airborne microorganisms, Methylobacterium spp. are capable of resisting desiccation to a certain degree and scavenging trace amounts of nitrogen and carbon which makes them well suited to survive in stressful environments66. Due to the lack of information on the residence time of these bacteria in the air, we can only speculate that the elevated cellular rRNA abundance in genus Methylobacterium compared to the average abundance from other genera could be attributed to their potential activities when airborne. However, this observation adds to a growing body of work on metabolic activity of microorganisms in the airborne phase18, 19. Thus, future investigations on the activity of environmentally-relevant bacterial species in airborne phase should consider Methylobacterium spp.
One advantage of analyzing 16S rRNA in combination with 16S rRNA gene sequences is the ability to identify the potentially active bacterial populations in environments of interest10, 11, 13, 19. In general, the 16S rRNA:16S rRNA gene ratios observed in this study (ranged from 0.07 to 16 with an average of 1.44 in summer; from 0.02 to 52 with an average of 2.00 in late winter/early spring) were comparable to the ratios (ranged from 0.002 to 122 with an average of 3.71) reported previously with airborne microbial community19. Besides, we also observed a pattern of the active microbial community that has been found in other environments13, 67–69 including the air19: bacterial taxa with higher 16S rRNA:16S rRNA gene ratios were also the rare members that exhibited lower relative 16S rRNA gene abundance than other taxa. However, current understanding of the function and ecological significance of those rare but potentially active microbes in the atmosphere is still limited19, and future research is warranted.
Our results showed that the absolute quantity of 16S rRNA in outdoor bioaerosols was generally almost two orders of magnitude higher than that of 16S rRNA gene (Figure 2a). The relatively higher abundance of rRNA greatly increases the sensitivity of detection method, especially for species that are rare in a particular environment10, allowing the reduction of air volume needed and, subsequently, the sampling time and effort needed to obtain sufficient sample size. Thus, 16S rRNA sequence analysis has a great potential to be applied in near realtime monitoring of potentially active bioaerosols, e.g. for public health monitoring and homeland surveillance purposes.
This study provides insight into a potential bias introduced by long-term air sampling stress on the accuracy of quantification and characterization of 16S rRNA in bioaerosols. We observed sampling device-dependent differences in 16S rRNA abundance for E. coli cells (laboratory experiments) and complex bacterial community (field experiments). Liquid impingement-based BioSampler recovered consistently less 16S rRNA than the filtration based Button sampler. The 16S rRNA sequences revealed different bacterial communities compared with the 16S rRNA gene-based results, and this difference depends on the sampling device. Overall, this study highlights the importance of selecting a sampling device when collecting and analyzing 16S rRNA in bioaerosols.
Supplementary Material
Highlights.
We investigated potential bias caused by air sampling stress on the measurement and characterization of 16S rRNA for bioaerosols in both laboratory and field experiments.
Liquid impingement-based BioSampler recovered consistently less 16S rRNA than the filtration based Button sampler.
The 16S rRNA sequences revealed a different bacterial community compared with 16S rRNA gene-based results, and this difference depended on the sampling device.
A number of bacterial taxa exhibited higher abundance in the 16S rRNA gene sequences than 16S rRNA sequences, which suggests potential metabolic activity of certain microbes in airborne phase.
Acknowledgments
The publication was supported by Grant R01-OH009783 “Advanced Sampler for Measuring Exposure to Biological Aerosols” from CDC/NIOSH, Grant IOS-1022254 “Air as an Active Bacterial Ecosystem” from the National Science Foundation (NSF), and Project 07160 funded by the New Jersey Agricultural Experiment Station (NJAES) at Rutgers, The State University of New Jersey. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of the CDC/NIOSH, the NSF, or the NJAES.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Woese CR. Bacterial evolution. Microbiol. Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van de Peer Y, Chapelle S, De Wachter R. A quantitative map of nucleotide substitution rates in bacterial rRNA. Nucleic Acids Res. 1996;24:3381–3391. doi: 10.1093/nar/24.17.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keer JT, Birch L. Molecular methods for the assessment of bacterial viability. J. Microbiol. Methods. 2003;53:175–183. doi: 10.1016/s0167-7012(03)00025-3. [DOI] [PubMed] [Google Scholar]
- 4.Josephson KL, Gerba CP, Pepper IL. Polymerase chain reaction detection of nonviable bacterial pathogens. Appl. Environ. Microbiol. 1993;59:3513–3515. doi: 10.1128/aem.59.10.3513-3515.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.England LS, Holmes SB, Trevors JT. Persistence of viruses and DNA in soil. World Journal of Microbiology & Biotechnology. 1998;14(2):163–169. [Google Scholar]
- 6.Cai P, Huang QY, Zhang XW. Interactions of DNA with clay minerals and soil colloidal particles and protection against degradation by DNase. Environ. Sci. Technol. 2006;40(9):2971–2976. doi: 10.1021/es0522985. [DOI] [PubMed] [Google Scholar]
- 7.Kerkhof L, Ward B. Comparison of nucleic acid hybridization and fluorometry for measurement of the relationship between RNA/DNA ratio and growth rate in a marine bacterium. Appl. Environ. Microbiol. 1993;59:1303–1309. doi: 10.1128/aem.59.5.1303-1309.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poulsen LK, Ballard G, Stahl DA. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl. Environ. Microbiol. 1993;59:1354–1360. doi: 10.1128/aem.59.5.1354-1360.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kemp PF, Lee S, LaRoche J. Estimating the growth rate of slowly growing marine bacteria from RNA content. Appl. Environ. Microbiol. 1993;59:2594–2601. doi: 10.1128/aem.59.8.2594-2601.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pitkänen T, Ryu H, Elk M, et al. Detection of fecal bacteria and source tracking identifiers in environmental waters using rRNA-based RT-qPCR and rDNA-based qPCR assays. Environ. Sci. Technol. 2013;47(23):13611–13620. doi: 10.1021/es403489b. [DOI] [PubMed] [Google Scholar]
- 11.Blazewicz SJ, Barnard RL, Daly RA, Firestone MK. Evaluating rRNA as an indicator of microbial activity in environmental communities: limitations and uses. ISME J. 2013;7:2061–2068. doi: 10.1038/ismej.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeAngelis KM, Wu CH, Beller HR, Brodie EL, et al. PCR amplification-independent methods for detection of microbial communities by the high-density microarray PhyloChip. Appl. Environ. Microbiol. 2011;77(18):6313–6322. doi: 10.1128/AEM.05262-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell BJ, Yu L, Heidelberg JF, Kirchman DL. Activity of abundant and rare bacteria in a coastal ocean. Proc. Natl. Acad. Sci. U.S.A. 2011;108(31):12776–12781. doi: 10.1073/pnas.1101405108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeAngelis KM, Silver W, Thompson AW, Firestone MK. Microbial communities acclimate to recurring changes in soil redox potential status. Environ. Microbiol. 2010;12(12):3137–3149. doi: 10.1111/j.1462-2920.2010.02286.x. [DOI] [PubMed] [Google Scholar]
- 15.Gentile G, Giuliano L, D'Auria G, Smedile F, Azzaro M, et al. Study of bacterial communities in Antarctic coastal waters by a combination of 16S rRNA and 16S rDNA sequencing. Environ. Microbiol. 2006;8(12):2150–2161. doi: 10.1111/j.1462-2920.2006.01097.x. [DOI] [PubMed] [Google Scholar]
- 16.Yarwood S, Brewer E, Yarwood R, Lajtha K, Myrold D. Soil microbe active community composition and capability of responding to litter addition after 12 years of no inputs. Appl. Environ. Microbiol. 2013;79(4):1385–1392. doi: 10.1128/AEM.03181-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez-Osorio AC, Williamson KS, Franklin MJ. Heterogeneous rpoS and rhlR mRNA levels and 16S rRNA/rDNA (rRNA gene) ratios within Pseudomonas aeruginosa biofilms sampled by laser capture microdissection. J Bacteriol. 2010;192(12):2991–3000. doi: 10.1128/JB.01598-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krumins V, Mainelis G, Kerkhof LJ, Fennell DE. Substrate-dependent rRNA production in an airborne bacterium. Environ. Sci. Technol. Letters. 2014;1(9):376–381. [Google Scholar]
- 19.Klein AM, Bohannan BJM, Jaffe DA, Levin DA, Green JL. Molecular evidence for metabolically active bacteria in the atmosphere. Front. Microbiol. 2016;7:772. doi: 10.3389/fmicb.2016.00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lighthart B. Minireview of the concentration variations found in the alfresco atmospheric bacterial populations. Aerobiologia. 2000;16:7–16. [Google Scholar]
- 21.Bauer H, Giebl H, Hitzenberger R, et al. Airborne bacteria as cloud condensation nuclei. J. Geophys. Res. 2003;108(D21):4658. [Google Scholar]
- 22.Mainelis G, Tabayoyong M. The effect of sampling time and the overall performance of portable microbial impactors. Aerosol Sci. Technol. 2010;44:75–82. [Google Scholar]
- 23.Zhen HJ, Han T, Fennell DE, Mainelis G. Release of free DNA by membrane-impaired bacterial aerosols due to aerosolization and air sampling. Appl. Environ. Microbiol. 2013;79(24):7780–7789. doi: 10.1128/AEM.02859-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Reponen T, Grinshpun SA, Gorny RI, Willeke K. Effect of sampling time and air humidity on the bioefficiency of filter samplers for bioaerosol collection. J. Aerosol Sci. 2001;32:661–674. [Google Scholar]
- 25.Deutscher MP. Degradation of stable RNA in bacteria. J. Biol. Chem. 2003;278:45041–45044. doi: 10.1074/jbc.R300031200. [DOI] [PubMed] [Google Scholar]
- 26.Hansen MC, Nielsen AK, Molin S, Hammer K, Kilstrup M. Changes in rRNA levels during stress invalidates results from mRNA blotting: Fluorescence in situ rRNA hybridization permits renormalization for estimation of cellular mRNA levels. J. Bacteriol. 2001;183(16):4747–4751. doi: 10.1128/JB.183.16.4747-4751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang CW, Chou FC. Assessment of bioaerosol sampling techniques for viable Legionella pneumophila by ethidium monoazide quantitative PCR. Aerosol Sci. Technol. 2011;45:343–351. [Google Scholar]
- 28.Rosset R, Julien J, Monier R. Ribonucleic acid composition of bacteria as a function of growth rate. J. Mol. Biol. 1966;18:308–320. doi: 10.1016/s0022-2836(66)80248-6. [DOI] [PubMed] [Google Scholar]
- 29.Hospodsky D, Yamamoto N, J P. Accuracy, precision, and method detection limits of quantitative PCR for airborne bacteria and fungi. Appl. Environ. Microbiol. 2010;76:7004–7012. doi: 10.1128/AEM.01240-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhen HJ, Krumins V, Fennell DE, Mainelis G. Development of a dual-internal-reference technique to improve accuracy when determining bacterial 16S rRNA:16S rRNA gene ratio with application to Escherichia coli liquid and aerosol samples. J. Microbiol. Methods. 2015;117:113–121. doi: 10.1016/j.mimet.2015.07.023. [DOI] [PubMed] [Google Scholar]
- 31.Aizenberg V, Grinshpun SA, Willeke K, Smith JP, Baron PA. Performance characteristics of the button personal inhalable aerosol sampler. Am. Ind. Hyg. Assoc. J. 2000;61:398–404. doi: 10.1080/15298660008984550. [DOI] [PubMed] [Google Scholar]
- 32.Han T, Mainelis G. Investigation of inherent and latent internal losses in liquid-based bioaerosol samplers. J. Aerosol Sci. 2012;45:58–68. [Google Scholar]
- 33.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 36.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007;73(5):1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, et al. Vegan: community ecology package. 2014 http://cran.r-project.org/web/packages/vegan/
- 40.Schimel J, Balser T, M W. Microbial stress-response physiology and its implicatiosn for ecosystem function. Ecology. 2007;88:1386–1394. doi: 10.1890/06-0219. [DOI] [PubMed] [Google Scholar]
- 41.Scherber CM, Schottel JL, Aksan A. Membrane phase behavior of Escherichia coli during desiccation, rehydration, and growth recovery. Biochim. Biophys. Acta. 2009;1788:2427–2435. doi: 10.1016/j.bbamem.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 42.Welsh DT, Herbert RA. Osmotically induced intracellular trehalose, but not glycine betaine accumulation promotes desiccation tolerance in Escherichia coli. FEMS Microbiol. Lett. 1999;174:57–63. doi: 10.1111/j.1574-6968.1999.tb13549.x. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Q, Yan T. Correlation of intracellular trehalose concentration with desiccation resistance of soil Escherichia coli populations. Appl. Environ. Microbiol. 2012;78(20):7407–7413. doi: 10.1128/AEM.01904-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Purusharth RI, Madhuri B, Ray MK. Exoribonuclease R in Pseudomonas syringae is essential for growth at low temperature and plays a novel role in the 3' end processing of 16 and 5 S ribosomal RNA. J. Biol. Chem. 2007;282(22):16267–16277. doi: 10.1074/jbc.M605588200. [DOI] [PubMed] [Google Scholar]
- 45.Cairrao F, Cruz A, Mori H, Arraiano CM. Cold shock induction of RNase R and its role in the maturation of the quality control mediator SsrA/tmRNA. Mol. Microbiol. 2003;50(4):1349–1360. doi: 10.1046/j.1365-2958.2003.03766.x. [DOI] [PubMed] [Google Scholar]
- 46.Chen CL, Deutscher MP. Elevation of RNase R in response to multiple stress conditions. J. Biol. Chem. 2005;280(41):34393–34396. doi: 10.1074/jbc.C500333200. [DOI] [PubMed] [Google Scholar]
- 47.Springorum AC, Clauß M, Hartung J. A temperature-controlled AGI-30 impinger for sampling of bioaerosols. Aerosol Sci. Technol. 2011;45:1231–1239. [Google Scholar]
- 48.Willeke K, Lin XJ, Grinshpun SA. Improved aerosol collection by combined impaction and centrifugal motion. Aerosol Sci. Technol. 1998;28:439–456. [Google Scholar]
- 49.Fierer N, Liu ZZ, Rodriguez-Hernandez M, Knight R, Henn M, Hernandez MT. Short-term temporal variability in airborne bacterial and fungal populations. Appl. Environ. Microbiol. 2008;74(1):200–207. doi: 10.1128/AEM.01467-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bowers RM, McCubbin IB, Hallar AG, Fierer N. Seasonal variability in airborne bacterial communities at a high-elevation site. Atmos. Environ. 2012;50:41–49. [Google Scholar]
- 51.Bowers RM, Sullivan AP, Costello EK, Collett JL, Jr, Knight R, Fierer N. Sources of bacteria in outdoor air across cities in the midwestern United States. Appl. Environ. Microbiol. 2011;77(18):6350–6356. doi: 10.1128/AEM.05498-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bowers RM, Clements N, Emerson JB, Wiedinmyer C, Hannigan MP, Fierer N. Seasonal variability in bacterial and fungal diversity of the near-surface atmosphere. Environ. Sci. Technol. 2013;47:12097–12106. doi: 10.1021/es402970s. [DOI] [PubMed] [Google Scholar]
- 53.Bowers RM, McLetchie S, Knight R, Fierer N. Spatial variability in airborne bacterial communities across land-use types and their relationship to the bacterial communities of potential source environments. ISME J. 2011;5:601–612. doi: 10.1038/ismej.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Corpe WA, Basile DV. Methanol-utilizing bacteria associated with green plants. Dev. Ind. Microbiol. 1982;23:483–494. [Google Scholar]
- 55.Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. The prokaryotes: A handbook on the biology of bacteria. In: Green PN, editor. Methylobacterium. Vol. 5. Spinger; New York, NY: 2006. pp. 257–265. [Google Scholar]
- 56.Kämpfer P, Martin K, McInroy JA, Glaeser SP. Novosphingobium gossypii sp. nov., isolated from Gossypium hirsutum. Int. J. Syst. Evol. Microbiol. 2015;65(9):2831–2837. doi: 10.1099/ijs.0.000339. [DOI] [PubMed] [Google Scholar]
- 57.Ceuppens S, Delbeke S, De Coninck D, Boussemaere J, Boon N, Uyttendaele M. Characterization of the bacterial community naturally present on commercially grown basil leaves: evaluation of sample preparation prior to culture-independent techniques. Int. J. Environ. Res. Public Health. 2015;12(8):10171–10197. doi: 10.3390/ijerph120810171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaewkla O, Franco CM. Rational approaches to improving the isolation of endophytic actinobacteria from Australian native trees. Microb. Ecol. 2013;65(2):384–93. doi: 10.1007/s00248-012-0113-z. [DOI] [PubMed] [Google Scholar]
- 59.Yamamura H, Ashizawa H, Nakagawa Y, Hamada M, Ishida Y, Otoguro M, Tamura T, Hayakawa M. Actinomycetospora rishiriensis sp. nov., isolated from a lichen. Int. J. Syst. Evol. Microbiol. 2011;61:2621–2625. doi: 10.1099/ijs.0.028753-0. [DOI] [PubMed] [Google Scholar]
- 60.Jones M, Harrison RM. The effects of meteorological factors on atmospheric bioaerosol concentrations - a review. Sci. Total Environ. 2004;326:151–180. doi: 10.1016/j.scitotenv.2003.11.021. [DOI] [PubMed] [Google Scholar]
- 61.Bowman JP, Cavanagh J, Austin JJ, Sanderson K. Novel Psychrobacter species from antarctic ornithogenic soils. Int. J. Syst. Evol. Microbiol. 1996;46:841–848. doi: 10.1099/00207713-46-4-841. [DOI] [PubMed] [Google Scholar]
- 62.Ehrlich R, Miller S, Walker RL. Effects of atmospheric humidity and temperature on the survival of airborne Flavobacterium. App. Microbio. 1970;20(6):884–887. doi: 10.1128/am.20.6.884-887.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xin YH, Liang ZH, Zhang DC, Liu HC, Zhang JL, Yu Y, Xu MS, Zhou PJ, Zhou YG. Flavobacterium tiangeerense sp. nov., a cold-living bacterium isolated from a glacier. Int. J. Syst. Evol. Microbiol. 2009;59:2773–2777. doi: 10.1099/ijs.0.007906-0. [DOI] [PubMed] [Google Scholar]
- 64.Hoisington A, Maestre JP, King MD, Siegel JA, Kinney KA. Impact of sampler selection on the characterization of the indoor microbiome via high-throughput sequencing. Building and Environment. 2014;80:274–282. [Google Scholar]
- 65.Lee Z-P, C B, Schmidt TM. rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea. Nucleic Acids Research. 2009;37:D489–D493. doi: 10.1093/nar/gkn689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Handley BA, Webster AJF. Some factors affecting the airborne survival of bacteria outdoors. J. Appl. Bacteriol. 1995;79:368–378. doi: 10.1111/j.1365-2672.1995.tb03150.x. [DOI] [PubMed] [Google Scholar]
- 67.Gremion F, Chatzinotas A, Harms H. Comparative 16S rDNA and 16S rRNA sequence analysis indicates that Actinobacteria might be a dominant part of the metabolically active bacteria in heavy metal-contaminated bulk and rhizosphere soil. Environ. Microbiol. 2003;(5):896–907. doi: 10.1046/j.1462-2920.2003.00484.x. [DOI] [PubMed] [Google Scholar]
- 68.Hugoni M, Taib N, Debroas D, Domaizon I, Jouan Dufournel I, Bronner G, et al. Structure of the rare archaeal biosphere and seasonal dynamics of active ecotypes in surface coastal waters. Proc. Natl. Acad. Sci. U.S.A. 2013;110:6004–6009. doi: 10.1073/pnas.1216863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilhelm L, Besemer K, Fasching C, Urich T, Singer GA, Quince C, et al. Rare but active taxa contribute to community dynamics of benthic biofilms in glacier-fed streams. Environ. Microbiol. 2014;16:2514–2524. doi: 10.1111/1462-2920.12392. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.