Figure 1. RT-PCR analysis of JCV late transcripts revealed an unexpected splice product.
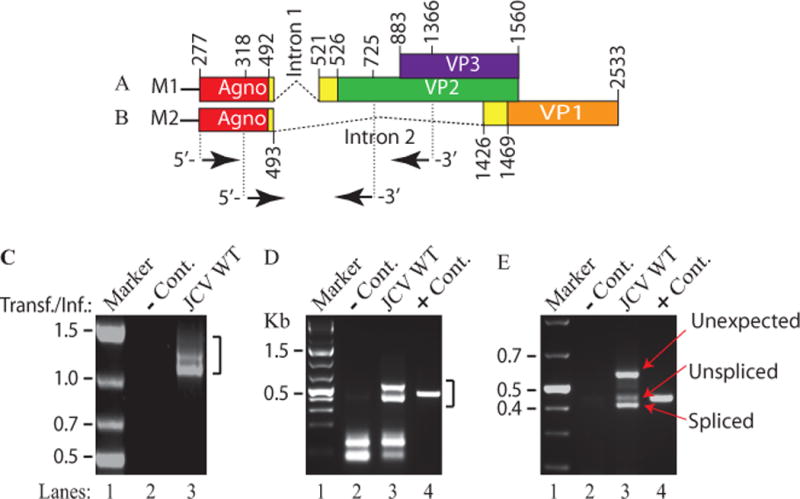
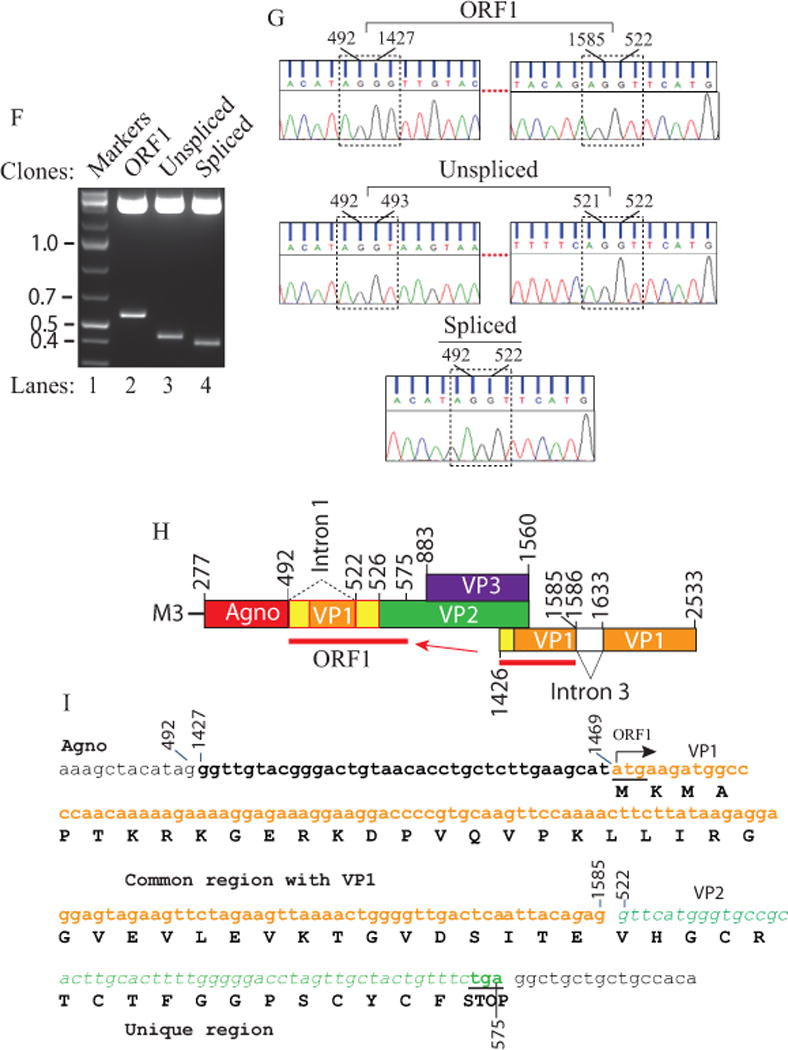
(A) Schematic representation of the JCV Mad-1 first major late transcript (M1) prior to splicing. The position of intron 1 between nucleotides 493 and 521 (JCV Mad-1 numbering) is indicated. (B) Schematic presentation of second major JCV Mad-1 late transcript (M2) prior to splicing. The position of intron 2 between nucleotides 493 and 1426 is indicated. (C) Agarose gel analysis of the RT-PCR products from the M1 transcript with specific primers (5′-JCV Mad-1 277-303 bp region and 3′-(JCV Mad-1, 1366-1346 bp). Total RNA was isolated from SVG-A cells transfected/infected with JCV Mad-1 WT at 15th day posttransfection/infection, subjected to RT-PCR reaction as described in Materials and Methods and analyzed on an agarose gel (1.5%). A bracket points the unresolved RT-PCR products. Transf./Inf.: Transfection/Infection. Total RNA from the untransfected SVG-A cells was also subjected to RT-PCR using the same primers as a negative control (− Cont.). (D) Re-amplification of the RT-PCR products from panel C by PCR using internal primers. The RT-PCR products from the panel C were re-amplified using the following internal primers: 5′-Primer: JCV Mad-1 (318-338) and 3′-primer: JCV Mad-1 (725-701) and resolved on an agarose gel (3 %). In lane 4, PCR-amplified JCV Mad-1 sequence was used as a positive control (+ Cont.) using the same internal primers. A bracket points to the unresolved bands. (E) Further resolution of the unresolved bands shown on panel D and subcloning them for sequencing. The resolved bands, labeled as “unexpected, unspliced and spliced” bands were gel-purified using QIAquick® gel extraction kit (Qiagen, catalog no. 28704), digested with HindIII and BamHI restriction enzymes and subcloned into the pcDNA3.1 (+) vector at HindIII and BamHI sites. Finally, the clones were sequenced commercially (Genewiz,http://www.genewiz.com). (F) Analysis of the cloned fragments by restriction enzyme digestion. The clones were digested with HindIII and BamHI enzymes and analyzed on a 3% agarose gel. (G) Partial representation of the DNA sequencing data of the cloned fragments at the splice junctions. The splice junctions were encased with dashed lines and the nucleotides were numbered according to the JCV Mad-1 strain numbering. (H) Schematic representation of a trans-spliced product (159 bp) of the 5′-end of the VP1 [Mad-1 (1426-1585) region] in place of “Intron 1” and creation of the ORF1 open reading frame. (I) Representation of the ORF1 sequence (159 bp) inserted in place of Intron 1. After insertion, a frame shift occurs within VP2, which is terminated with stop codon at nucleotide position 575. The common region of ORF1 (41 aa long) with VP1 is colored orange and the unique region is colored green (17 aa long).
