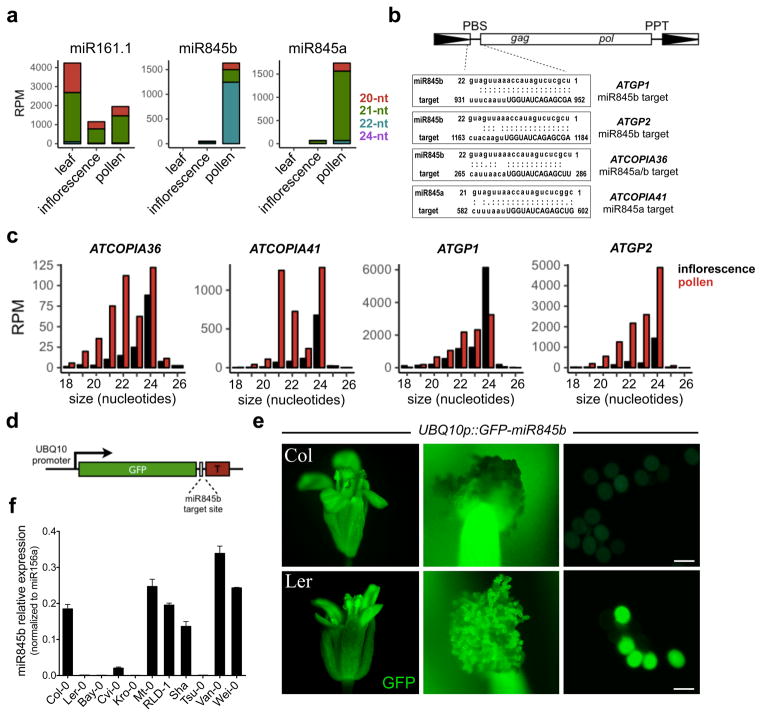
Figure 1. miR845 family is expressed in pollen and targets retrotransposons.
(a) miR845a and miR845b are preferentially expressed in mature pollen, consistent with complete absence in leaves and low levels in inflorescence tissue. (b) miR845 targets the PBS (uppercase) of retrotransposons where tRNAs bind to initiate reverse transcription. Predicted targets include Gypsy and Copia elements. The full list of targets is presented in Supplementary Table 1. (c) miR845 targets accumulate high levels of secondary 21- and 22-nt easiRNA in pollen. RPM, reads per million. (d) GFP sensor construct includes 3′UTR with a miR845b target site and driven by the UBIQUITIN10 (UBQ10) promoter. (e) Strong GFP fluorescence was detected in floral organs of 8 independent transgenic lines, but not in wild type Col-0 pollen. The same reporter is not silenced in Ler-0 pollen, where miR845b is not expressed. Scale bars represent 30 μm. (f) The MIR845 haplotype in Ler-0 is also found in other Arabidopsis accessions such as Bay-0, Kro-0 and Tsu-0 that produce low levels of miR845b, while Col-like accessions express high levels of miR845b in pollen. Bars represent mean Ct values of miR845b levels as measured by quantitative RT-PCR (qRT-PCR) and normalized to the levels of miR156a (n=2 technical replicates). Error bars represent range.