Fig. 7. Comparisons of S. lebanonensis and D. melanogaster Grk protein and gene sequences reveal significant homology.
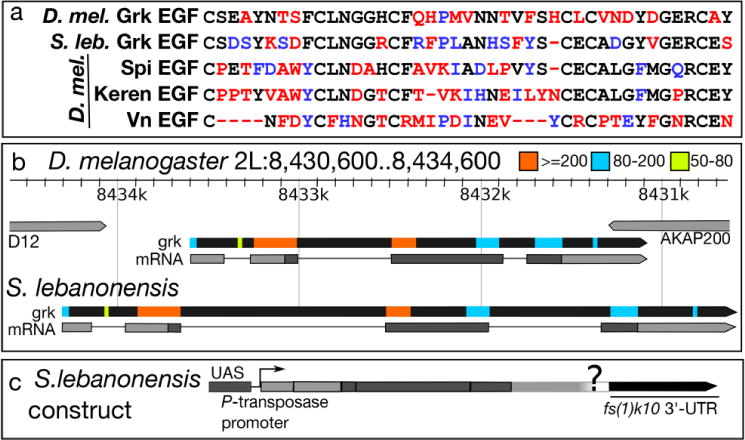
(a) Amino acid alignment of the EGF domains of four EGF ligands in D. melanogaster and the EGF domain from the predicted Grk protein from S. lebanonensis; similarity with D. melanogaster Grk is shown in descending order. Black letters represent identical amino acids; blue letters represent amino acids with similar chemical properties; red letters represent amino acids that differ chemically. (b) Top, D. melanogaster grk gene locus and transcript (grk-RA); see (http://flybase.org/reports/FBgn0001137.html) for details. S. lebanonensis grk gene structure below. Gene structures are to scale. Colored boxes indicate homologous regions with the associated bit scores. (c) Schematic diagram of the construct used to test GrkSl activity in D. melanogaster.
