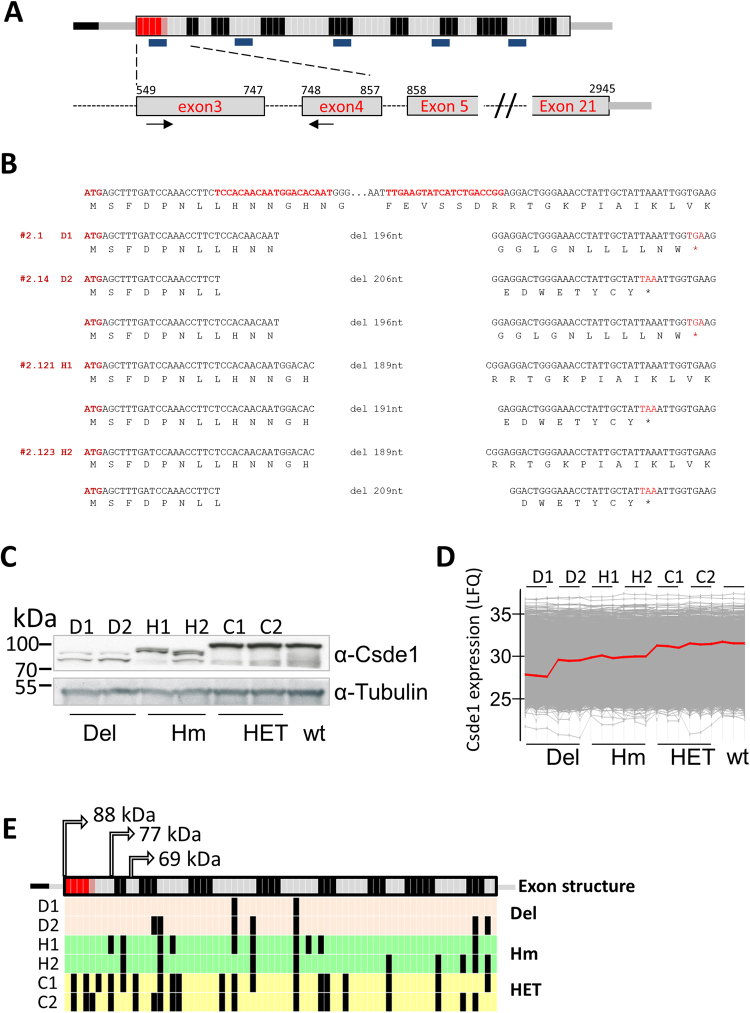
Figure 3.
Deletion of the 1st cold shock domain of Csde1 using Crispr/Cas9. (A) Cartoon of the Csde1 transcript. Grey and black represent subsequent exons. Small squares represent individual tryptic peptides (exons and peptides in arbitrary size). Tryptic peptides were assigned to the exon that contributes most. The methionine start codon locates at the start of exon 3. The position of five cold shock domains is indicated by short bars below the transcript. Numbers on the zoom in on exons 3–5 indicate the nucleotide position of NM_144901.4. Guide RNAs are indicated with arrows, the red tryptic fragments are affected by the deletion. (B) The top sequence shows the guide RNAs in red, in their sequence context, and the amino acids below the codons. Below the sequence that reveals the precise deletion for both alleles in 4 clones, only 1 allele was detected in clone D1 (#2.1). (C) Western blot stained for Csde1 and tubulin as loading control. The membrane was cut between Csde1 and tubulin and staining performed on each separately. D1 (2.1) and D2 (2.14) represent out-of-frame deletions (Del), H1 (2.121) and H2 (2.123) in-frame deletions of the 1st cold shock domain (Hm). HET are control clones that harbor 1 deleted (out-of-frame) allele and 1 wt allele. Raw image scans are available as Supplementary Figure S6. (D) Mass spectrometry analysis with MaxQuant and Perseus yields a cartoon in which the protein expression of all proteins is indicated as LFQ by a grey line. Csde1 expression is indicated with a red line. (E) For Csde1, we identified 33 unique tryptic peptides. Black lines indicate tryptic fragments that were detected by mass spectrometry. Alternative AUG start codons that may explain truncated proteins are indicated by arrows, the size of the encoded protein in kDa.