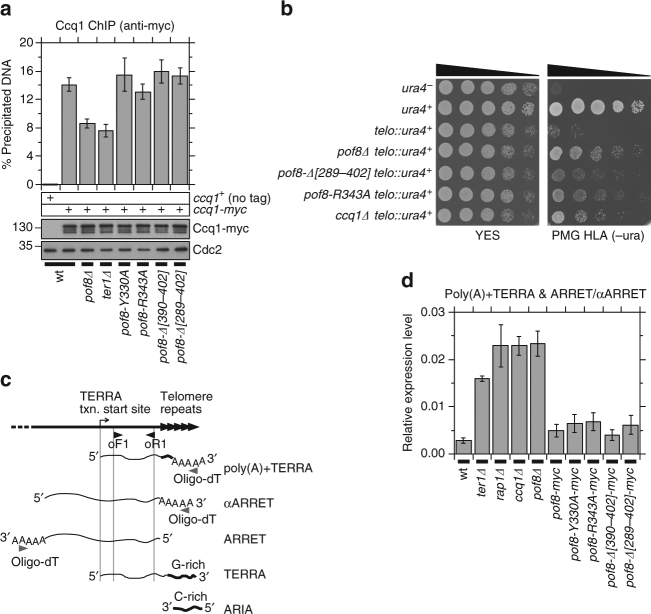
Fig. 7.
Pof8 protein is required to repress transcription at telomere/sub-telomere regions. a Quantitative real-time PCR ChIP analysis to monitor Ccq1 association with telomeres for indicated genetic backgrounds. Expression levels of myc-tagged Ccq1 used in ChIP assays were monitored by western blot analysis. Anti-Cdc2 blots served as loading control. Molecular weight (kDa) of size markers are also indicated. b Transcriptional silencing at telomeres is disrupted in pof8∆ cells. Wild-type and pof8 mutant cells carrying the ura4+ marker gene at the chromosome 2 L telomere48 were serially diluted and spotted on YES (no selection) or PMG HLA (-ura) plates. c A schematic diagram indicating various lncRNA species expressed at fission yeast telomere/sub-telomere regions26, 49. Locations of primers used in RT reaction (oligo-dT) and subsequent quantitative PCR analysis (oF1 and oR1) to monitor poly(A)-tailed lncRNAs are indicated. d Pof8 protein, but not its xRRM domain is required to repress transcription of poly(A)-tailed lncRNAs at telomere/sub-telomere regions. Expression levels of telomeric transcripts were normalized to his1+ mRNA. For plots in a and d, error bars correspond to SEM from at least five independent experiments. Raw data and statistical analysis are available in Supplementary Data 1