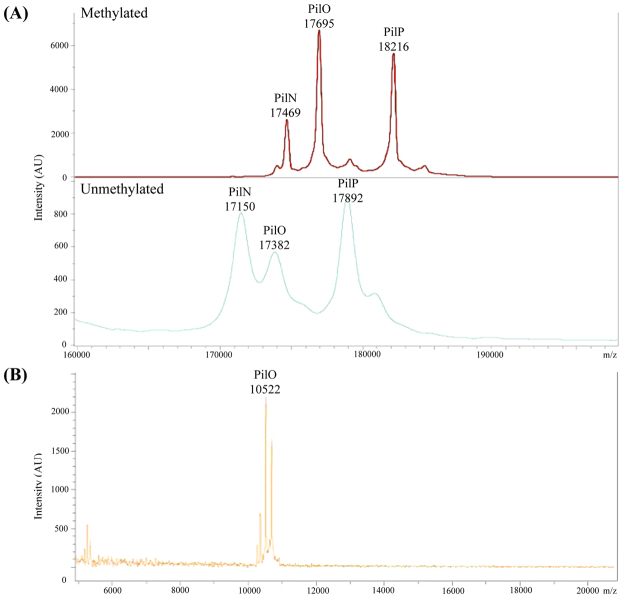
Figure 1.
Mass spectrometry (MALDI-TOF/TOF) results indicate successful methylation of the PilNΔ44/PilOΔ51/PilPΔ18 complex but only PilO fragments were present in the crystals. (A) Methylated (top) and unmethylated (bottom) samples of 1 mg/mL PilNΔ44/PilOΔ51/PilPΔ18 was analyzed on a Bruker UltrafleXtreme linear detector in positive ion mode. The change in the molecular weight of PilN, PilO and PilP between the methylated and unmethylated samples were 319 Da, 313 Da, and 324 Da, respectively. All three protein fragments have 11 lysine residues, which could be methylated. A single dimethyl-Lys group adds 28 Da. (B) Two crystals were washed and resuspended in buffer and was analyzed on a Bruker UltrafleXtreme linear detector in positive ion mode. This weight corresponds to a 110–206 fragment of PilO (approximate molecular weight of 10,560 Da). Remaining peaks–were confirmed to be various PilO fragments. The numbers above the peaks are the molecular weights shown in Da. The chromatogram shows the ion intensities (in arbitrary units) according to mass-to-charge (m/z) ratio.