Figure 1.
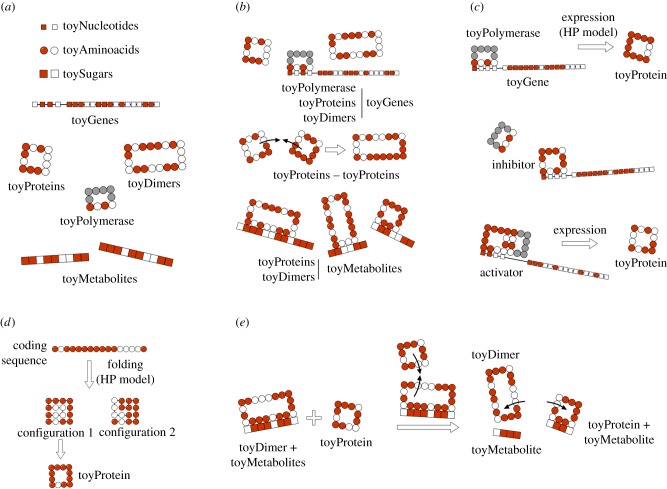
Brief overview of toyLIFE. (a) The three basic building blocks of toyLIFE are toyNucleotides, toyAminoacids and toySugars. They can be hydrophobic (H, white) or polar (P, red), and their random polymers constitute toyGenes, toyProteins and toyMetabolites. The toyPolymerase is a special polymer that will have specific regulatory functions. These polymers will interact between each other following an extension of the HP model, for which we have chosen the interaction energies EHH = − 2, EHP = − 0.3 and EPP = 0 [17]. (b) Possible interactions between pairs of toyLIFE elements. toyGenes interact through their promoter region with toyProteins (including the toyPolymerase and toyDimers); toyProteins can bind to form toyDimers, and interact with the toyPolymerase when bound to a promoter; both toyProteins and toyDimers can bind a toyMetabolite at arbitrary regions along its sequence. (c) A toyGene is expressed (translated) when the toyPolymerase binds to its promoter region. The sequence of Ps and Hs of the toyProtein will be exactly the same as that of the toyGene coding region. If a toyProtein binds to the promoter region of a toyGene with a lower energy than the toyPolymerase does, it will displace the latter, and the toyGene will not be expressed. This toyProtein acts as an inhibitor. The toyPolymerase does not bind to every promoter region. Thus, not all toyGenes are expressed constitutively. However, some toyProteins will be able to bind to these promoter regions. If, once bound to the promoter, they bind to the toyPolymerase with their rightmost side, the toyGene will be expressed, and these toyProteins act as activators. (d) toyProteins fold on a 4 × 4 lattice, following a self-avoiding walk (SAW). For each binary sequence of length 16, we fold it into every SAW and compute its folding energy, following the HP model. Then we choose the SAW that yields the minimum folding energy. (e) Metabolism in toyLIFE. A toyDimer is bound to a toyMetabolite when a new toyProtein comes in. If the new toyProtein binds to one of the two units of the toyDimer, forming a new toyDimer energetically more stable than the old one, the two toyProteins will unbind and break the toyMetabolite up into two pieces. We say that the toyMetabolite has been catabolized. (Online version in colour.)