Figure 1. Seed dependent RNAi off-target effects (sOTE).
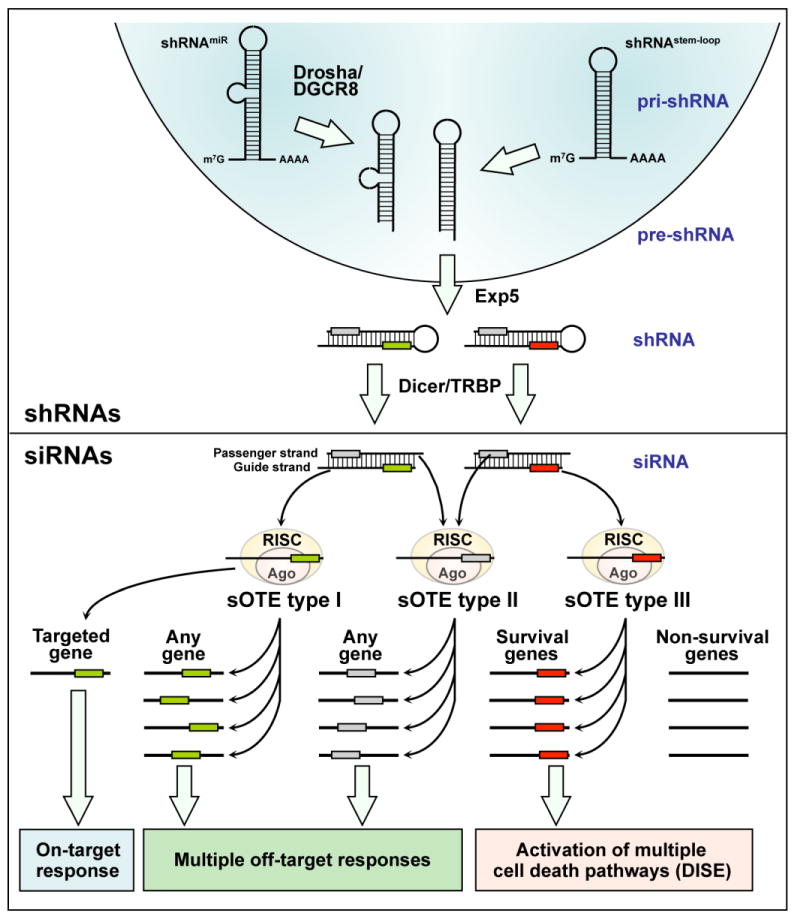
shRNAs, introduced most often in the form of a lentiviral vector and inserted into the genome, are transcribed by either RNA Pol II or III. They can either be simple stem loop shRNAs (shRNAstem-loop) or miRNA-based next generation shRNAs (shRNAmiR), which require Drosha/DGCR8 processing before they are exported into the cytosol by exportin 5 (Exp5). Each shRNA contains a sense/passenger strand and a complimentary antisense/guide strand. Dicer/TRBP trims these duplexes, thereby removing the loop region. Both trimmed shRNAs and exogenous and endogenous siRNAs are loaded into the RNA induced silencing complex (RISC) with their Argonaute proteins (Ago). Similar to miRNAs, positions 2-7 (the seed sequence) of the loaded guide strand greatly determines what target mRNA is silenced through complementarity of the seed sequence, mostly to the 3′UTR of the targeted mRNAs. In addition to the silencing of the mRNA that the siRNA was designed to target, siRNAs show three types of seed dependent off target effects (sOTE). sOTE type I: The guide strand gets properly loaded into the RISC but its seed sequence (green box) targets other genes expressed in treated cells that have complementarity to the siRNA seed. Because this type of sOTE can affect any type of gene, the resulting cellular responses are multiple. sOTE type II: the passenger strand of the siRNA, rather than the intended guide strand, is loaded into the RISC. Position 2-7/8 at the 5′ end of the passenger (now guide) strand (grey box) becomes the seed sequence, and targeting of mRNAs that show complementarity to the passenger strand seed sequence again results in multiple cellular responses. sOTE type III: Certain seed sequences in the guide strand of specialized RNAs (red box) preferentially target seed matches present in the 3′UTR of survival genes and to a lesser extent non-survival genes. This sOTE results in the cell death of the treated cells called DISE.
