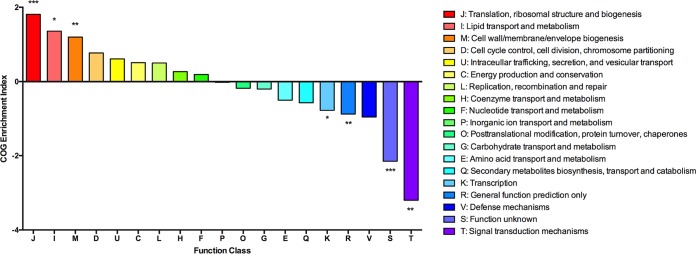
FIG 2 .
Essential genes by Clusters of Orthologous Groups (COG) classification. Essential and compromised genes that are annotated with known functions are plotted according to the COG enrichment index. This index is calculated as the percentage of the essential genome made up of a COG divided by the percentage of the whole genome made up by the same COG. The log2 fold enrichment is displayed, and significant differences were calculated using the two-tailed Fisher’s exact test. If the COG enrichment index for a given COG is >0, it implies that a greater proportion of genes within that COG are essential than would be expected and vice versa for a COG enrichment index of <0. Log2 COG enrichment index values that are significantly different from expected are indicated by asterisks as follows: *, P < 0.05; **, P < 0.01;***, P < 0.001.