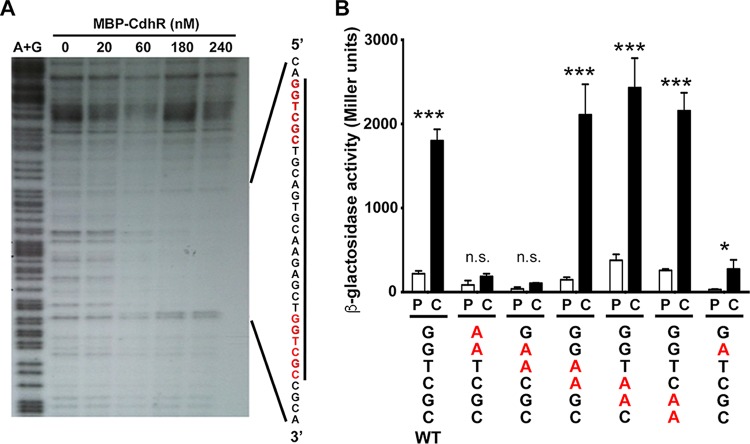
FIG 3 .
The CdhR binding site and key residues for CdhR-dependent induction. (A) A DNase I footprinting assay was performed by taking the caiX UAS end labeled with 32P and adding increasing concentrations of MBP-CdhR, followed by DNase I treatment and nondenaturing 5% polyacrylamide TBE gel. The first lane of the gel is the A+G sequencing ladder, and the nanomolar concentration of MBP-CdhR is marked. (B) The caiX enhancer site was mutated by changing two bases at a time (in red and underlined) in the caiX distal binding site to adenosines and fused to lacZ. The P. aeruginosa PA14 wild type carrying each of the plasmids was grown in MOPS with 20 mM pyruvate at 20 µg·ml−1, with or without 1 mM carnitine for 4 h, and then β-galactosidase activity was reported as Miller units. Error bars represent standard deviations from three biological replicates, and results are representative of three independent experiments. Data were analyzed using a two-way analysis of variance (ANOVA) with a Sidak’s multiple-comparison posttest comparing each mutant’s pyruvate to carnitine. Abbreviations: P, pyruvate; C, carnitine; n.s., not significant; *, P < 0.05; ***, P < 0.001.