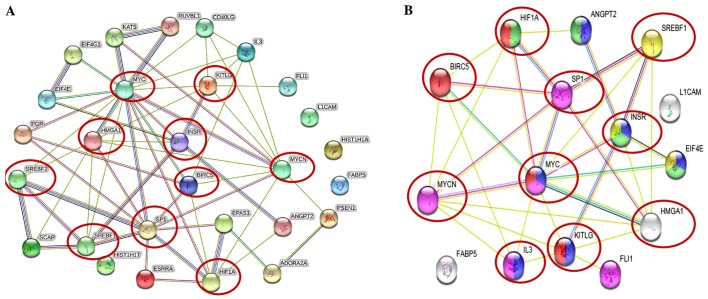
Figure 2.
Tumorigenic proteins significantly upregulated in the IMR-32 cells mapped to the MYCN proto-oncogene, bHLH transcription factor (MYCN) pathway. (A) STRING pathway analysis revealed interactions between molecules of activated upstream regulators in IMR-32 cells as predicted by IPA. Of importance are the various proteins with hubs (multiple edges, red circles), including Sp1 transcription factor (SP1), MYC, MYCN, hypoxia-inducible factor-1α (HIF-1α), high mobility group protein A1 (HMGA1), INSR, KIT ligand (KITLG), sterol regulatory element binding transcription factor 2 (SREBF2) and SREBF1. (B) STRING pathway analysis also focused on the strong interactions between the activated upstream regulators which HMGA1, MYCN and BIRC5 mapped to. Of particular interest are the hubs, including MYCN, MYC, SP1, HIF-1α, SREBF1, BIRC5, HMGA1 and INSR that affiliated with one or more of our validated targets.