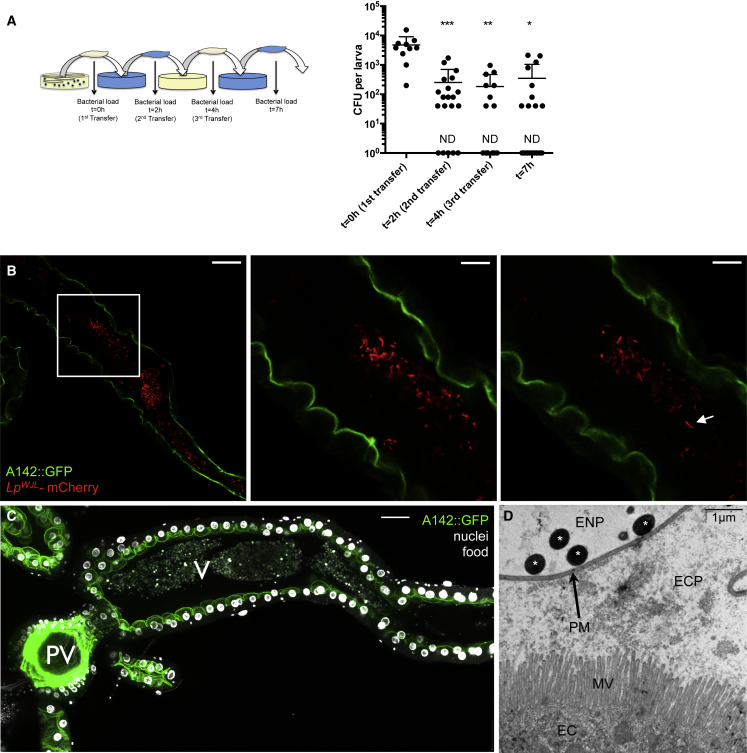
Figure 2.
L. plantarum Transits in the Endoperitrophic Space with the Food Bolus
(A) Evolution of the larval bacterial load after repeated transfers on axenic food. Left panel: experimental setup. Right panel: bacterial load quantification. To plot all data points on a log scale, a value of “1” was attributed to samples with no detectable CFUs and these have been marked “ND” (not detected). Asterisks represent a statistically significant difference with the initial bacterial burden (t = 0 hr).
(B) Ingested bacteria occupy the central part of the gut lumen. Anterior midgut of an A142::GFP larva fed on food containing LpWJL expressing mCherry. GFP localizes to the brush border and thus the apical side of the enterocytes. Individual mCherry-expressing bacteria or pairs of bacilli (arrow) can be seen in the lumen. The samples were mounted unfixed. Single confocal sections are shown. Images for the center and right panels were taken at higher magnification (zoom 3×) than for the left panels (white square) and they are distinct sections of one z stack. Scale bars, 50 μm (left), 16.67 μm (center and right).
(C) A142::GFP gut fixed and stained with DAPI to mark nuclei. Autofluorescence highlights the food bolus. PV, the proventriculus; V, the ventriculus. Scale bars, 50 μm (B and C). Note the apparent gap between the larval tissue (enterocyte epithelium) and the mass of fluorescent bacteria or food, both seem to occupy the endoperitrophic space.
(D) Transmission electron microscopy of anterior midgut transversal sections of 6DAEL LpWJL-mono-associated larvae reared on PYD. White asterisks, bacteria; PM, peritrophic matrix; ECP, ectoperitrophic space; ENP, endoperitrophic space; MV, microvilli; EC, enterocyte. Scale bar, 1 μm.
Asterisks represent a statistically significant difference with the initial bacterial burden (t = 0 hr): ∗∗∗0.0001 < p < 0.001, ∗∗0.001 < p < 0.01, ∗p < 0.05.