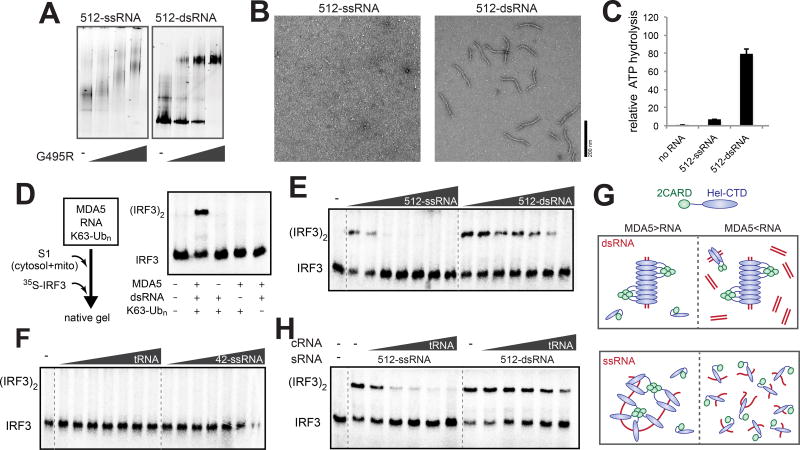
Figure 2. RNA-rich cellular environment necessitates MDA5 filament assembly on dsRNA for signaling.
(A–C) Native gel shift assay (A), representative electron micrographs (B) and ATPase activity (C) of G495R in complex with 512-ssRNA and 512-dsRNA (see Methods for the RNA sequence). 2CARD deletion mutant (Δ2CARD) of MDA5, which is both necessary and sufficient for RNA binding, was used for all EM, native gel shift assays and ATPase assays in this study. In all comparisons, same mass concentrations (2.5, 0.6 and 0.4 ng/µl for native gel, EM and ATPase assays, respectively) of RNAs were used.
(D) Schematic of the cell-free IRF3 dimerization assay using S1 extract from 293T cells. Right: native gel showing 35S-IRF3 dimerization in the presence and absence of MDA5, dsRNA and K63-Ubn.
(E and F) IRF3 stimulatory activity of G495R in complex with an increasing concentration (0.5–50 ng/µl) of 512-ssRNA and 512-dsRNA (E), and yeast tRNA and 42-ssRNA (F).
(G) A model to explain the observed difference between dsRNA and ssRNA in stimulating the MDA5 signaling activity. On dsRNA (upper panel), cooperative filament formation allows proximity-induced oligomerization of 2CARD regardless of the level of RNA. On ssRNA (lower panel), however, receptor oligomerization occurs only when RNA is present at substoichiometric concentrations (left), but not in excess (right).
(H) IRF3 stimulatory activity of G495R in complex with stimulatory RNA (sRNA), 512-ssRNA and 512-dsRNA (both at 0.5 ng/µl), in the presence of an increasing concentration (0.5–50 ng/µl) of non-stimulatory, competitor tRNA (cRNA).
See also Figure S2.