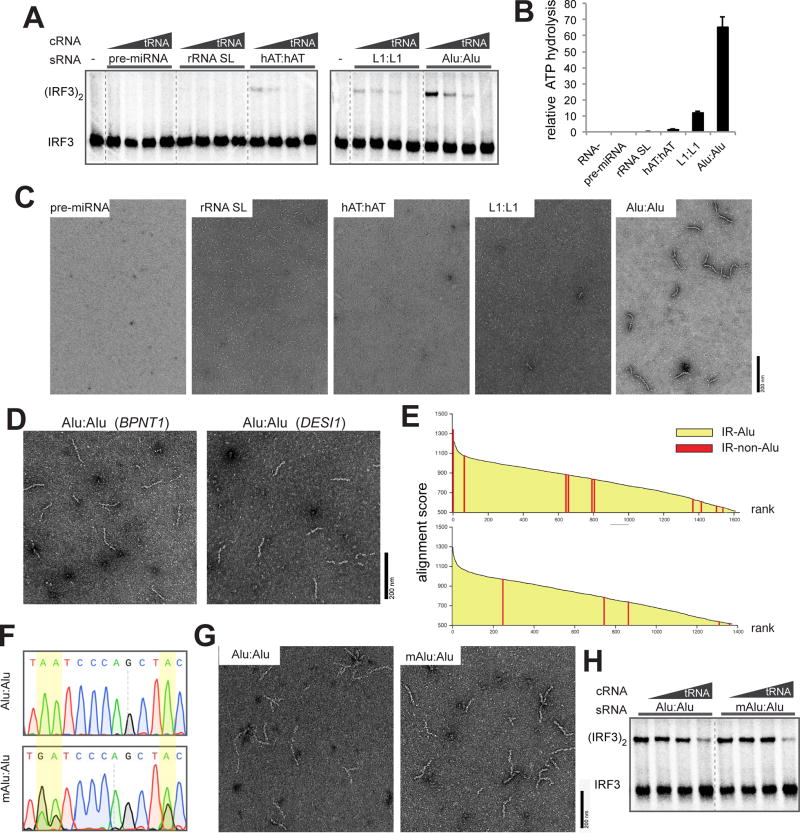
Figure 4. Alu:Alu hybrids, unlike other cellular dsRNA, can robustly stimulate MDA5 filament formation and signaling.
(A) IRF3 stimulatory activity of G495R in complex with stimulatory RNAs (sRNA, 0.5 ng/µl) that represent five types of cellular dsRNAs. The activity was measured with an increasing concentration of competitor tRNA (cRNA, 0–8 ng/µl).
(B and C) ATPase activity (B) and representative electron micrographs (C) of G495R in complex with cellular dsRNAs (0.4 ng/µl for ATPase assay and 0.6 ng/µl for EM) used in (A). Data are represented as mean ± SD (n=3) for (B).
(D) Representative electron micrographs of G495R filaments formed on Alu:Alu hybrids from BPNT1 and DESI1 3’UTRs.
(E) Alignment scores between adjacent inverted repeats. Pairs of inverted repeats were ranked by the alignment scores (Y-axis), and listed in the descending order (X-axis). Two scoring systems were used for sequence alignment: default from the program Exonerate (top) and modified parameters with higher penalty for gaps (bottom). Red bars represent pairs of inverted repeats that are not Alu elements; there are 11 (top) and 5 (bottom) non-Alu pairs out of ~1500 total inverted repeat pairs. All the remaining pairs (yellow) are IR-Alus.
(F) Sequencing chromatograms of Alu:Alu hybrids (NICN1 3’UTR) before and after in vitro A-to-I modification by ADAR1. Yellow highlights indicate the positions of the modification. Note that an A-to-I-modified base is reverse transcribed as G.
(G and H) Representative electron micrographs (G) and IRF3 stimulatory activity (H) of G495R in complex with Alu:Alu hybrids (NICN1 3’UTR) before and after A-to-I modification.
See also Figure S4.