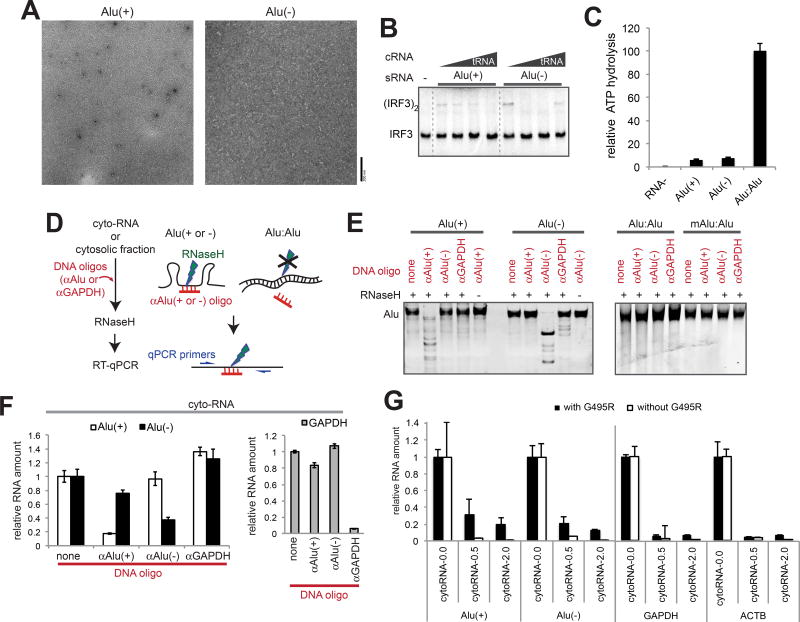
Figure 5. Paired Alus, not unpaired Alus, stimulate GOF MDA5 and are abundant in cytosol.
(A–C) Representative electron micrographs (A), IRF3 stimulatory activity (B), and ATPase activity (C) of G495R in complex with the sense (+) or antisense (−) strand of Alu from the NICN1 3’UTR.
(D) Schematic of the RNase H-based method to selectively cleave unpaired, but not paired Alu RNAs.
(E) Gel analysis of the RNase H assay. In vitro transcribed Alu RNAs (from NICN1 3’UTR) were subjected to the RNase H assay as described in (D). An oligo targeting GAPDH (αGAPDH) was used for negative controls.
(F) Quantitation of Alu:Alu hybrids in cytosolic RNA. The RNase H assay in (D) was performed using purified cytosolic RNA from 293T cells, and remaining Alu(+) and Alu(−) were quantitated relative to the spike-in control.
(G) The levels of Alu(+), Alu(−), GAPDH and ACTB (right) relative to the spike-in control before and after the RNase A protection assay. CytoRNA-0.0, −0.5 and −2.0 indicate RNAs recovered after digestion with 0.0, 0.5 and 2.0 ng/µl RNase A in the presence or absence of G495R.
Data represent mean ± SD (n=3) for (C), (F) and (G). See also Figure S5.