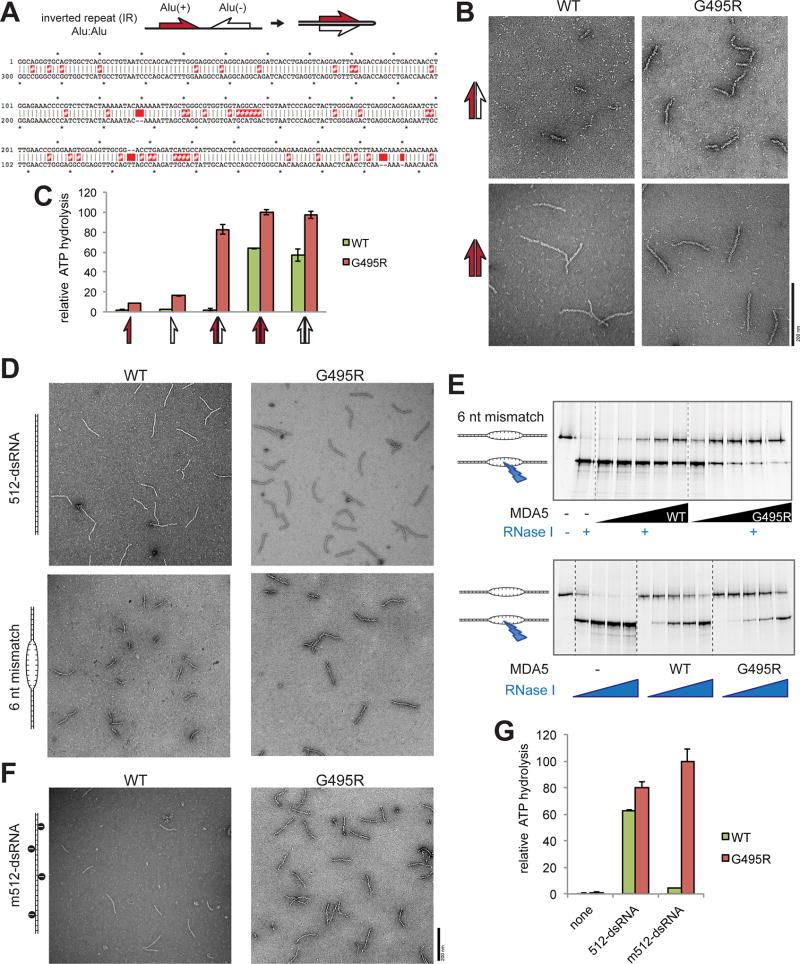
Figure 6. WT MDA5 is sensitive to dsRNA structural irregularities and is thus inefficient in recognizing an imperfect duplex of Alu:Alu hybrid.
(A) Schematic of Alu:Alu hybrids formed by IR-Alus in NICN1 3’UTR. Red and white half arrows indicate sense (+) and antisense (−) Alus, respectively. Below is the sequence alignment of Alu(+) (top strand) and the reverse complement of Alu(−) (bottom strand). Red # and space indicate mismatch and bulge, respectively.
(B) Representative electron micrographs of WT and G495R in complex with the naturally occurring Alu:Alu hybrids from NICN1 3’UTR (red:white arrow, top) or with an artificial perfect duplex formed by Alu(+) and its reverse complement (red:red arrow, bottom).
(C) ATPase activity of WT and G495R when bound by unpaired or paired Alus from NICN1 3’UTR. Arrows are as defined in (A and B).
(D) Representative electron micrographs of WT and G495R in complex with 512 bp dsRNA with or without 6 nt mismatch at the center.
(E) RNase I footprinting assay to examine the occupancy of the 6 nt mismatched site by WT or G495R molecules. The RNase I sensitivity was examined with an increasing concentration of MDA5 (top) or RNase I (bottom). The saturating concentration (1 µM) of MDA5 was used in the bottom to compare WT and G495R independent of their differential affinities for dsRNA.
(F and G) Representative electron micrographs (F) and ATPase activity (G) of WT and G495R in complex with A-to-I modified 512-dsRNA. Data are mean ± SD (n=3) for (C and H).
See also Figure S6.