Figure 7. Unmodified Alu:Alu hybrids activate wild-type MDA5 under the ADAR1-deficiency.
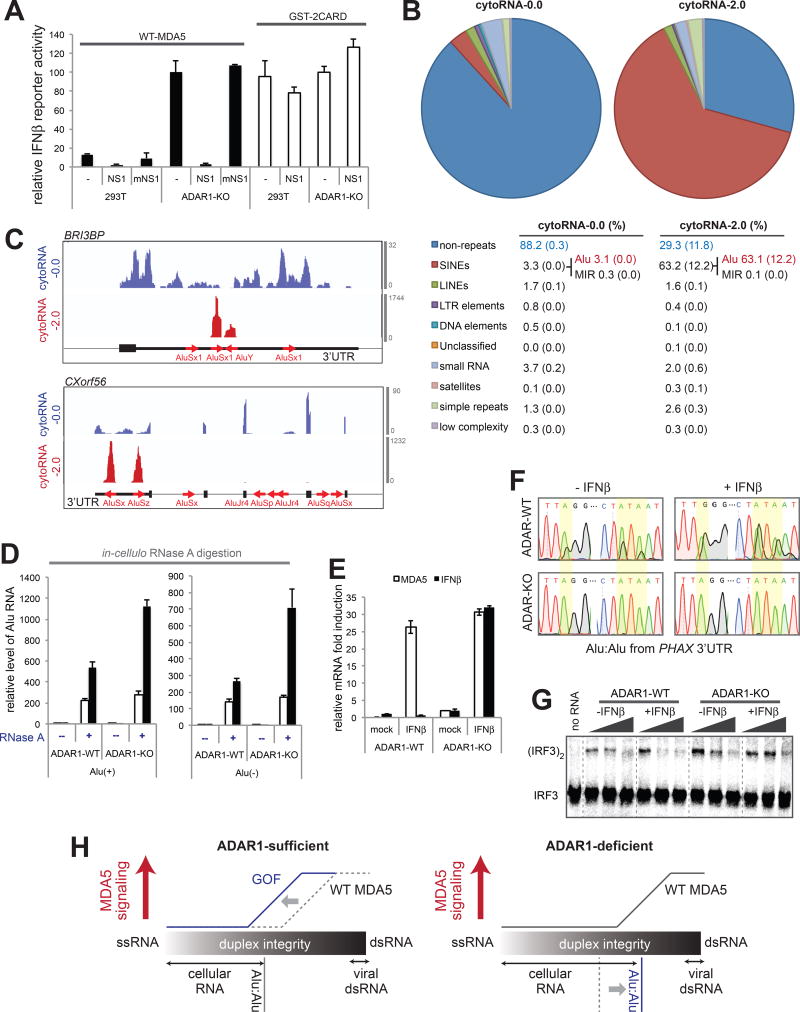
(A) The IFNβ reporter activity of WT MDA5 and GST-2CARD in the presence or absence of NS1/mNS1 in ADAR1-WT and -KO cells.
(B) RNA-seq followed by RepeatMasker analysis of cytoRNA-0.0 and cytoRNA-2.0, which were generated by WT MDA5-protected digestion of ADAR1-KO cytosolic RNAs. The numbers represent averages (standard deviations in parenthesis) of the two independent biological repeats.
(C) Distribution of sequencing reads of cytoRNA-0.0 and cytoRNA-2.0. Two representative genes (BRI3BP and CXorf56) from the top enriched genes are shown as in Figure 3G.
(D) The level of Alu RNA relative to GAPDH after in-cellulo RNase A protection assay. WT MDA5Δ2CARD (or empty vector, EV) was ectopically expressed in ADAR1-WT or -KO cells and RNase A was introduced into the cells using SLO pores as in Figure 3J.
(E) The level of IFNβ or MDA5 (IFIH) mRNA induction upon treatment with the IFNβ protein in ADAR1-WT and -KO cells.
(F) Sequencing chromatograms of representative IR-Alus (in PHAX 3’UTR) from ADAR1-WT and -KO cytosol before and after (24 hr post) IFNβ treatment.
(G) IRF3 stimulatory activity of WT MDA5 in complex with increasing concentration of cytosolic RNA (0.5–15 ng/µl) from ADAR1-WT and -KO cells before and after IFNβ treatment.
(H) A model of how Alu:Alu hybrids activate GOF MDA5 in ADAR1-sufficient cells (left) and WT MDA5 in ADAR1-deficient cells (right). In ADAR1-sufficient cells, WT MDA5 does not recognize cellular RNAs due to its sensitivity to structural irregularities caused by mismatches, bulges and A-to-I modifications. GOF mutations, however, make MDA5 less sensitive to such dsRNA structural irregularities, allowing recognition of imperfect duplexes such as Alu:Alu hybrids. In ADAR1-deficient cells, on the other hand, the lack of A-to-I modification increases the structural integrity of Alu:Alu hybrids, allowing aberrant recognition by WT MDA5.
Data represent mean ± SD (n=3) for (A), (D) and (E). See also Figure S7.