Abstract
MYO18A is a divergent member of the myosin family characterized by the presence of an amino-terminal PDZ domain. MYO18A has been found in a few different complexes involved in intracellular transport processes. MYO18A is found in a complex with LURAP1 and MRCK that functions in retrograde treadmilling of actin. It also has been found in a complex with PAK2, βPIX, and GIT1, functioning to transport that protein complex from focal adhesions to the leading edge. Finally, a high proportion of MYO18A is found in complex with GOLPH3 at the trans Golgi, where it functions to promote vesicle budding for Golgi-to-plasma membrane trafficking. Interestingly, MYO18A has been implicated as a cancer driver, as have other components of the GOLPH3 pathway. It remains uncertain as to whether or not MYO18A has intrinsic motor activity. While many questions remain, MYO18A is a fascinatingly unique myosin that is essential in higher organisms.
Keywords: MYO18A, myosin, GOLPH3, Golgi, cell migration, cancer
1. Introduction
MYO18A is an unconventional myosin that has been implicated in multiple cellular processes. It has roles in retrograde treadmilling of actin and in the function of focal adhesions. It also plays an important role at the Golgi as part of the machinery for vesicle exit for Golgi-to-plasma membrane trafficking. There remain many questions about its function and controversies in the literature that have yet to be resolved. Nevertheless, it is clear that MYO18A is an interesting protein whose study promises to provide insight into a range of biology.
2. Discovery of MYO18A as a Novel PDZ-Containing Myosin
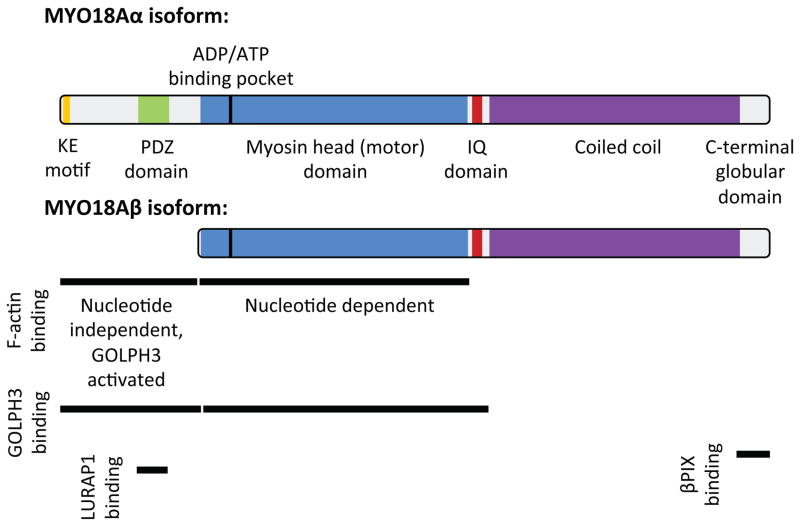
MYO18A was first identified in 2000 by Obinata and colleagues in a screen for genes that are up-regulated in stromal cell lines with increased capacity to support hematopoietic stem cells (presumably via secretion of paracrine growth factors) (Furusawa et al., 2000). Their further characterization of MYO18A (referred to as MysPDZ) reported ubiquitous expression in all tissues, detection of multiple splice forms, and subcellular localization of both the endogenous protein and exogenously expressed, tagged protein at the ER and Golgi (Furusawa et al., 2000; Mori et al., 2003, 2005). Analysis of MYO18A domain structure revealed a core myosin-homology region, closest in homology to myosin II but fairly divergent, an extended coiled-coil region at the C-terminus, and an extended N-terminus (Figure 1). The N-terminal portion of the protein was recognized to contain a Lys-Glu (KE) repeat region. In addition, unique among myosins, the MYO18A N-terminal portion includes a PDZ domain, which is a critical defining feature of MYO18A across species. PDZ domains typically enable protein-protein interactions often by binding to a PDZ motif found at the C-terminus of some proteins. However, there also are examples of PDZ domain interactions with internal protein sequences (Harris and Lim, 2001) and with lipids (Zimmermann, 2006). PDZ domains are frequently observed in proteins involved in intracellular signal transduction pathways. Thus, even early on it seemed that MYO18A may be involved in signaling. Bolstering this idea, unbiased analysis of protein phosphorylation in myeloblastic cells in response to macrophage colony stimulating factor (CSF-1) identified prominent tyrosine phosphorylation of MYO18A (Cross et al., 2004). Indeed, subsequent unbiased proteomic analyses have identified abundant phosphorylation of MYO18A on Ser, Thr, and Tyr, in addition to other post-translational modifications such as lysine ubiquitination, acetylation, and methylation (Hornbeck et al., 2015). The function, if any, of these modifications remains largely unknown.
Fig. 1.
Protein domain structure and protein interaction sites of MYO18A. See text for details and references.
3. A Role for MYO18A in Retrograde Actin Treadmilling
Tan et al. identified MYO18A in a complex with Myotonic dystrophy kinase-related CDC42-binding kinase (MRCK) and Leucine-rich adaptor protein 1 (LURAP1, also known as LRAP35a) (Tan et al., 2008). Unbiased experiments to identify proteins that co-immunoprecipate with LURAP1 identified MYO18A. They found that LURAP1 bridges from MRCK to MYO18A, and that MRCK is capable of phosphorylating MYO18A. Functional experiments revealed that overexpression of MRCK, LURAP1, or MYO18A resulted in an increase in cellular protrusions. Further experiments indicated this phenotype to be due to a role of the complex in retrograde flow of the actomyosin network in the cell lamella and the center of the cell. Finally, these authors provided evidence that the MRCK-LURAP1-MYO18A complex is required for normal cell migration.
4. A Role for MYO18A in Linking Focal Adhesions to Membrane Ruffles
MYO18A has also been found to play a role in the turnover of focal adhesions and their function in regulating membrane ruffles. In a pair of studies, Hsu et al. found that MYO18A interacts with the PAK2/βPIX/GIT1 complex (Hsu et al., 2010, 2014). Interestingly, the C-terminus of βPIX does contain a consensus PDZ motif, but the data suggest that the βPIX C-terminus interacts with the C-terminal globular domain of MYO18A, rather than the PDZ domain (Hsu et al., 2014). They found that depletion of MYO18A does not alter the formation of the PAK2/βPIX/GIT1 complex, however it alters the localization of the complex. Under normal conditions in epithelial cells, the PAK2/βPIX/GIT1 complex cycles (or traffics) between the leading edge (membrane ruffles and lamellipodia), focal adhesions, and the cytoplasm. Depletion of MYO18A, or disruption of the MYO18A-βPIX interaction, results in the accumulation of the PAK2/βPIX/GIT1 complex in focal adhesions. A consequence of this focal adhesion accumulation of the PAK2/βPIX/GIT1 complex is decreased focal adhesion turnover, with a resultant increase in both the size and number of focal adhesions. This occurs together with a reduction in active membrane ruffles and is associated with decreased cell migration. These data suggest a model in which MYO18A transports the PAK2/βPIX/GIT1 complex from mature focal adhesions to membrane ruffles, and that this drives cell migration.
5. A Role for MYO18A in Lung Surfactant Function
Chroneos and colleagues report a role for MYO18A as a receptor for lung surfactant protein A, SP-A (Sever-Chroneos et al., 2011; Yang et al., 2005, 2015). They provide evidence of an interaction between MYO18A and SP-A, and propose that there exists a cryptic transmembrane domain in MYO18A that results in the extracellular localization of the motor and C-terminal domains. Certainly, this would provide an unexpected twist to MYO18A function. However, important questions remain as how a protein that is generally observed to be cytosolic, lacks a definitive export signal sequence, and lacks a definitive transmembrane domain could act as a receptor for molecules found in the extracellular space. Additional studies will be needed to understand whether MYO18A truly plays this intriguing role.
6. MYO18A Has a Key Role in Golgi Function Via an Interaction with GOLPH3
6.1 The PI4P/GOLPH3/MYO18A/F-actin complex
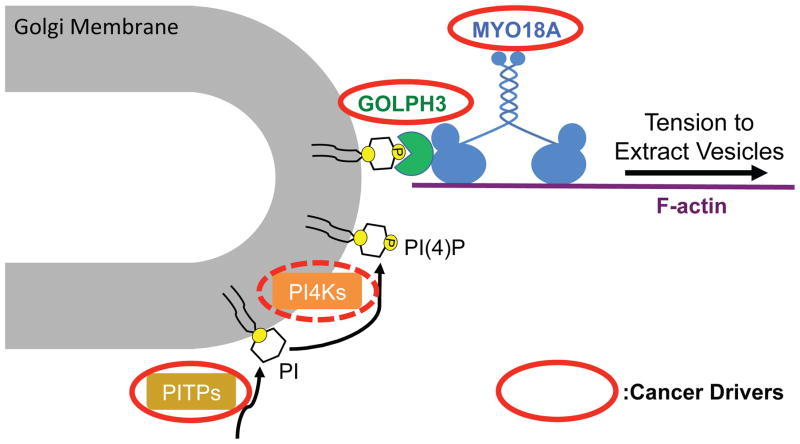
GOLPH3 is an abundant protein that is highly localized to the cytosolic face of the trans Golgi (Figure 2; Bell et al., 2001; Dippold et al., 2009; Wu et al., 2000). In a screen for phosphoinositide binding proteins we identified GOLPH3 as a novel PI4P-binding protein (Dippold et al., 2009). PI4P is highly abundant at the trans Golgi (Godi et al., 1999, 2004). GOLPH3 binding to trans Golgi PI4P results in localization of GOLPH3 to the trans Golgi (Dippold et al., 2009). We found that siRNA knockdown of GOLPH3 results in compaction of the Golgi and arrest of Golgi-to-plasma membrane trafficking (Dippold et al., 2009), mimicking the effect of depolymerization of F-actin (Dippold et al., 2009; Egea et al., 2006; Hirschberg et al., 1998; Lázaro-Diéguez et al., 2007). The similarity of the phenotypes suggested a link between GOLPH3 and F-actin. Thus, it was gratifying that unbiased detection of GOLPH3 interacting proteins by immunoprecipitation/mass spectrometry (IP/MS) identified MYO18A (Dippold et al., 2009). Further experiments indicated that quantitative IP of endogenous GOLPH3 would co-IP as much as two-thirds of endogenous MYO18A, indicating a robust interaction between the endogenous proteins (Dippold et al., 2009; Farber-Katz et al., 2014; Ng et al., 2013). Furthermore, the purified proteins interact directly with each other in vitro (Dippold et al., 2009; Farber-Katz et al., 2014; Taft et al., 2013). In agreement with Mori, et al. (Mori et al., 2005) we found that MYO18A localizes in part to the Golgi, and further, the Golgi localization of MYO18A is dependent on GOLPH3 (Dippold et al., 2009). Our data indicate that the PI4P/GOLPH3/MYO18A/F-actin complex functions to pull on the trans Golgi membrane to extract vesicles for forward trafficking to the plasma membrane (Bishé et al., 2012; Dippold et al., 2009; Farber-Katz et al., 2014; Ng et al., 2013; Xing et al., 2016).
Fig. 2.
MYO18A is a critical component of the Golgi GOLPH3 pathway. See text for details and references.
6.2 The GOLPH3/MYO18A Interaction is Regulated
The interaction between GOLPH3 and MYO18A undergoes physiological regulation as part of the cellular response to DNA damage (Farber-Katz et al., 2014). In response to agents that cause double-stranded DNA breaks, activated DNA-PK directly phosphorylates GOLPH3 on Thr143 and Thr148. This phosphorylated form of GOLPH3 interacts more tightly with MYO18A, as observed both with endogenous proteins in cells, and with bacterially expressed, purified proteins in vitro (Farber-Katz et al., 2014). Moreover, this enhanced interaction between GOLPH3 and MYO18A is responsible for fragmentation of the Golgi that occurs in response to DNA damage.
6.3 A Role for the Golgi GOLPH3/MYO18A Complex in Cell migration
MYO18A also plays a role in cell migration as a key component of the GOLPH3 pathway at the Golgi. Overexpression of GOLPH3 has been shown by several groups to promote increased cell migration (Isaji et al., 2014; Tokuda et al., 2014; Xing et al., 2016). It has long been known that a key initiating step in cell migration is reorientation or polarization of the Golgi towards the leading edge of the cell (Kupfer et al., 1982; Millarte and Farhan, 2012). Our lab showed that the GOLPH3 pathway drives cell migration by promoting Golgi reorientation towards the leading edge and by increasing directional trafficking of cargoes towards the leading edge (Xing et al., 2016). Furthermore, we found that all components of the Golgi GOLPH3 pathway (PI4P, GOLPH3, MYO18A and F-actin) are required for reorientation of the Golgi and efficient cell migration.
6.4 Loss of MYO18A Phenocopies Loss of GOLPH3
Consistent with MYO18A acting in concert with GOLPH3, siRNA knockdown of MYO18A phenocopies knockdown of GOLPH3. For example, knockdown of GOLPH3 (or depletion of PI4P, or depolymerization of F-actin) causes the Golgi ribbon to collapse into a compact ball as observed by light microscopy and causes the normally flattened trans Golgi cisternae to round-up as observed by electron microscopy (Dippold et al., 2009; Ng et al., 2013). Knockdown of GOLPH3 also causes arrest of Golgi-to-plasma membrane trafficking (as assessed by unbiased 35S-methionine pulse-chase to measure overall secretion, quantitative assessment of trafficking of ts045-VSVG-EGFP, or secretion of hepatitis C virus from infected cells). Knockdown of MYO18A completely recapitulates these phenotypes (Bishé et al., 2012; Dippold et al., 2009; Ng et al., 2013). Knockdown of GOLPH3 also prevents DNA damage-induced dispersal of the Golgi and results in enhanced cell death following DNA damage. Again, knockdown of MYO18A reproduces these phenotypes (Farber-Katz et al., 2014). In cell culture wound healing experiments to assess cell migration, knockdown of GOLPH3 interferes with reorientation of the Golgi toward the wound edge and inhibits cell migration. Knockdown of MYO18A results in the same defects in Golgi reorientation and impaired cell migration (Xing et al., 2016). Moreover, epistasis experiments reveal that the ability of overexpressed GOLPH3 to accelerate Golgi reorientation toward the wound edge is dependent on MYO18A and F-actin (Xing et al., 2016). The observation that knockdown of MYO18A thoroughly phenocopies knockdown of GOLPH3 argues strongly that they function in concert. Moreover, the fact that knockdown of MYO18A is epistatic to overexpression of GOLPH3 indicates that the function of GOLPH3 depends on MYO18A.
6.5 GOLPH3L Reveals the Importance of the GOLPH3/MYO18A Interaction
GOLPH3L is a paralog of GOLPH3, found in vertebrates. It serves as an experiment of nature revealing the importance of the interaction between GOLPH3 and MYO18A. GOLPH3L is highly similar to GOLPH3, and like GOLPH3 binds tightly and specifically to PI4P, and thus localizes to the Golgi (Ng et al., 2013). However, the Golgi phenotype due to knockdown or overexpression of GOLPH3L is opposite to that of GOLPH3. Knockdown of GOLPH3L causes Golgi expansion, while overexpression of GOLPH3L causes Golgi compaction. The explanation for the disparity between the effect of perturbing GOLPH3L versus GOLPH3 became clear with the discovery that while GOLPH3 interacts tightly with MYO18A, GOLPH3L does not. Furthermore, epistasis analysis indicates that GOLPH3L’s ability to influence the Golgi is dependent on both GOLPH3 and MYO18A. Thus, GOLPH3L acts as an endogenous dominant-negative inhibitor of the GOLPH3/MYO18A complex.
7. A Role for MYO18A in Cancer
7.1 MYO18A Drives Cancers
To identify new cancer driver genes, Sanchez-Garcia et al. developed a new algorithm (Helios) to mine The Cancer Genome Atlas (TCGA) breast cancer database (Sanchez-Garcia et al., 2014). This analysis identified several well established oncogenes (such as MYC, ERBB2/Neu, and cyclin D1) as cancer drivers. Along with the well characterized cancer driving genes, they identified several novel putative cancer drivers, including MYO18A. Further validating their Helios analysis system, several of these previously unknown cancer drivers were validated as bona fide oncogenes in cell transformation models. The robust validation of Helios provides some assurance that their identification of MYO18A as a cancer driver is also true. However, direct experimental evidence is lacking. Still, Makowska et al. find that MYO18A is overexpressed in metastatic prostate cancer cell lines (Makowska et al., 2015). In addition, the MYO18A gene has been found in fusions with the PDGFR, FGFR, and MLL genes in cancers (Sheng et al., 2017; Ussowicz et al., 2012; Walz et al., 2005, 2009). While PDGFR, FGFR, and MLL are known cancer drivers, it remains possible that the MYO18A portion of these fusions is also driving these cancers.
7.2 Multiple Components of the GOLPH3 Pathway Are Cancer Drivers
Notably, other components of the GOLPH3 pathway are also cancer drivers. GOLPH3 itself is an oncogene that is amplified and/or overexpressed in many cancers (Buschman et al., 2015; Scott et al., 2009). PITPNC1 is a member of the family of phosphatidylinositol transfer proteins (PITPs) and is a cancer driver (Halberg et al., 2016; Png et al., 2011). Strikingly, the phenotypes due to overexpression of PITPNC1 are dependent on GOLPH3, indicating that PITPNC1 acts via the GOLPH3/MYO18A pathway (Halberg et al., 2016). The Golgi localized PI-4-kinases, PI4KIIα (Li et al., 2010) and PI4KIIIβ (Curtis et al., 2012) have also been proposed to be cancer drivers. Taken together, the data suggest that the components of the GOLPH3 pathway, including MYO18A, drive oncogenesis through their function at the Golgi (Buschman et al., 2015; Farber-Katz et al., 2014; Xing et al., 2016).
7.3 MYO18A Drives Migration of Prostate Cancer Cells
A role for MYO18A in cell migration and actin organization was reported in studies performed in prostate cancer cells (Makowska et al., 2015). This work showed that MYO18A expression (along with several other myosins) is increased in invasive prostate cancer cell lines, supporting the idea that MYO18A plays a role in oncogeneses and/or metastasis. Furthermore, depletion of MYO18A from PC-3 prostate cancer cells results in altered actin cytoskeleton (particularly non-muscle myosin 2A fibers) and impaired cell migration and invasion. Specifically, they found that MYO18A depletion has no effect on cell migratory speed, but instead impairs persistent movement, and thus productive migration.
Although the study in prostate cancer did not propose a mechanism, it is notable that three mechanisms have already been proposed for MYO18A’s contribution to cell migration. Tan et al. suggested that MYO18A’s role in retrograde treadmilling of actin contributes to cell migration (Tan et al., 2008). Hsu et al. proposed that MYO18A’s role in trafficking of the PAK2/βPIX/GIT1 complex from focal adhesions to membrane ruffles is important for cell migration (Hsu et al., 2010, 2014). Finally, Xing et al. (our work) indicated a role for MYO18A in Golgi re-orientation and directional trafficking towards the leading edge, which are known to be important for cell migration (Kupfer et al., 1982; Millarte and Farhan, 2012; Xing et al., 2016). These proposed mechanisms are not mutually exclusive, and thus it is certainly possible that all three contribute to the requirement for MYO18A for normal and invasive cell migration.
8. Is MYO18A a Motor?
The myosin family of proteins is known by their ability to bind actin and to use the energy from hydrolysis of ATP to translocate on actin filaments. Since MYO18A is clearly a member of the myosin family by homology, it is natural to wonder whether it likewise acts as a motor on actin. Unexpectedly, when Isogawa et al. characterized the interaction of human MYO18A with F-actin in vitro, they were unable to detect an interaction between the motor domain and F-actin, even after depletion of ATP, which for other myosins results in a tight rigor complex (Isogawa et al., 2005). They did find that a region of the N-terminus of MYO18A, between the KE and PDZ domains did bind F-actin, although this binding was unaffected by depletion of ATP. Together with the observation that key residues found in other myosins are not conserved in MYO18A, the authors proposed that MYO18A may not act as a motor.
Subsequent studies of MYO18A in vitro have cast further suspicion on the ability of MYO18A to act as a motor. Characterization of fruit fly MYO18A observed no ability to bind or hydrolyze ATP (Guzik-Lendrum et al., 2011). The same group examined mouse MYO18A and found that while it could bind to myosin light chains, it bound ATP only weakly, hydrolyzing it quite inefficiently (Guzik-Lendrum et al., 2013). They also observed only weak binding to actin filaments that was unregulated by nucleotide or addition of GOLPH3. Because of the weak actin binding, in this study the authors were unable to see binding of MYO18A to actin filaments as observed by the formation of barbed arrows by electron microscopy.
Taft et al. examined human MYO18A in vitro, and observed two actin binding sites per monomer, yielding four sites per MYO18A dimer (Taft et al., 2013). The motor domain was found to bind equally well to ATP and ADP, and to bind actin better in the presence of ADP compared to in the presence of ATP or in the absence of nucleotide. Furthermore, the KE-PDZ region of the N-terminus was also found to bind to actin (as was previously observed by Isogawa et al.), and although this binding was not regulated by nucleotides, it was enhanced five-fold by the addition of GOLPH3 protein. Taft et al, were readily able to observe the formation of barbed arrows on actin filaments indicating efficient decoration by MYO18A.
The differences observed by these different groups could be due to species differences (fruit fly vs. mouse vs. human). However, it seems more likely to represent experimental differences, including the use of different fragments of MYO18A by each group. Perhaps more significant, Isogawa and Taft used bacterially expressed MYO18A, which is likely to be devoid of regulatory post-translational modifications. Guzik-Lendrum et al., in their studies chose to use protein produced using the baculovirus/Sf9 system, which may bear random post-translational modifications. Differences in post-translational modifications may significantly affect MYO18A activity. Nevertheless, every group reported detecting little or no ATP hydrolysis by MYO18A, raising doubts as to its ability to act as a motor.
Unlike the experiments performed in vitro, experiments performed in live cells suggest that the putative ATP catalytic site, as identified by homology to other myosins, is functionally important. Tan et al. observed that overexpression of the putative ATPase-defective MYO18A mutant G520D/K521E behaved similarly to depletion of MRCK or MYO18A, causing a marked reduction in the actomyosin network in the lamellar and subnuclear regions (Tan et al., 2008). This suggested that the ATPase activity is required for MYO18A function in cells, and that the ATPase-defective mutant behaves as a dominant-negative.
In Dippold et al. we used knockdown/rescue experiments to test the function of MYO18A at the Golgi (Dippold et al., 2009). siRNA knockdown of MYO18A in human cells resulted in Golgi compaction. Normal Golgi morphology could be rescued by expression of murine wild-type MYO18A (which is resistant to the human-targeted siRNA), but not by expression of the putative ATPase-defective mutant, G520S/K521A. The fact that subtle mutation in the putative ATP catalytic site renders MYO18A nonfunctional in cells for both Tan et al. and Dippold et al. strongly raises suspicion that MYO18A catalyzes nucleotide hydrolysis in vivo.
The difference in conclusions offered by the biochemical experiments performed in vitro and the functional experiments performed in live cells could be explained in a number of ways. It is possible that the point mutations used in the cell-based experiments alter MYO18A function in some other unforeseen manner that has nothing to do with nucleotide hydrolysis, and, in fact, MYO18A is not a motor. It is noteworthy that all of the cell-based functions for MYO18A, such as actomyosin treadmilling, trafficking of the PAK2/βPIX/GIT1 complex, or Golgi vesicle budding in concert with GOLPH3, could involve additional motor proteins that interact either directly or indirectly with MYO18A to generate the proposed forces. Indeed, MYO18A has been found to co-assemble with non-muscle myosin 2 both in vitro and in live cells (Billington et al., 2015). It is certainly possible that MYO18A depends on co-assembled non-muscle myosin 2 to generate force.
On the other hand, it is important to maintain a healthy skepticism when interpreting the lack of an activity observed in experiments performed in vitro (and, more generally, for any negative data). There are many ways for an experiment to fail, including the use of inappropriate buffer conditions, the lack of critical cofactors, missing partner proteins, or inappropriate post-translational modifications. Indeed, GOLPH3 and MYO18A are both heavily post-translationally modified, and at least some of these modifications are functionally important (Bell et al., 2001; Farber-Katz et al., 2014; Hornbeck et al., 2015; Wu et al., 2000). As a cautionary tale, the literature provides many examples of kinase family proteins that were proclaimed to be pseudokinases for many years based on lack of activity in vitro, and lack of key amino acid residues that were supposedly critical for function. Examples include CASK (Mukherjee et al., 2008), Erb3/HER3 (Shi et al., 2010), Wnk1 (Xu et al., 2000) and Haspin (Eswaran et al., 2009) that eventually were proven to be active kinases both in vitro and in vivo. At present the data are not sufficiently compelling to determine whether MYO18A does or does not function as a motor. Clearly, additional data are needed.
9. Function of MYO18A in Model Organisms
9.1 Mhcl is the Drosophila MYO18A Ortholog
Two studies have examined the function of the MYO18A ortholog found in Drosophila melanogaster, which is called Mhcl (myosin heavy chain-like). Ivanov et al. performed a large scale RNAi screen to identify genes that are required for normal embryonic development of the nervous system (Ivanov et al., 2004). They screened RNAi targeted to 3,314 genes, and found 43 that interfere with normal neural development, including Mhcl. Injection of RNAi specific for Mhcl resulted in dramatic loss of central and peripheral neurons in fly embryos.
Bonn et al. investigated the expression pattern of Mhcl mRNA, noting that the levels increased sharply in myoblast founder cells just before and during the process of myoblast fusion that gives rise to mature myotubes (Bonn et al., 2013). They observed Mhcl protein adjacent to the sarcomeric Z-discs in mature myotubes. In developing myoblasts and myotubes they observed GFP-Mhcl in myoblast founder cells at the membrane where fusion occurs with fusion competent myoblast cells. From this and other data they proposed a role for Mhcl in myoblast fusion. Interestingly, myoblast fusion is known to be dependent on Golgi-derived vesicles (Doberstein et al., 1997; Lu et al., 2001), raising the possibility that this function in myoblasts actually reflects a role for Mhcl at the Golgi.
These authors then generated a genetic deletion of the Mhcl locus (deleting Mhcl plus five adjacent genes) (Bonn et al., 2013). This homozygous deletion of Mhcl was uniformly lethal by the first larval stage. The authors did not observe abnormalities of muscle, or any other abnormalities in the embryos, oddly in disagreement with the results of Ivanov et al. (Ivanov et al., 2004). Bonn et al. proposed that in Drosophila other myosins are able to compensate for the loss of Mhcl (MYO18A).
It is perhaps worth observing that MYO18A appears late in evolution, and there is no ortholog of MYO18A in simpler eukaryotes such as yeast. Nevertheless, GOLPH3 is conserved back through yeast. In the budding yeast Saccharomyces cerevisiae, Golgi-to-plasma membrane trafficking is dependent on PI4P (Audhya et al., 2000; Hama et al., 1999; Walch-Solimena and Novick, 1999) and the yeast type V myosin (MYO2) (Govindan et al., 1995; Johnston et al., 1991; Santiago-Tirado et al., 2011; Schott et al., 1999). In fact, the yeast type V myosin has been reported to indirectly interact with PI4P via an unknown adaptor protein (Santiago-Tirado et al., 2011). GOLPH3 (VPS74 in yeast) is an obvious candidate, since in mammals it does serve to bind PI4P and bridge to a myosin. It seems likely that prior to the evolution of MYO18A that other myosins functioned with GOLPH3, so perhaps it is not surprising that in early organisms that have a MYO18A ortholog (e.g., Drosophila) that some of its function is dispensable.
9.2 MYO18A in Zebrafish
In zebrafish, gene duplication gave rise to two MYO18A genes, myo18aa and myo18ab. In a pair of publications, Cao et al. have studied the function of the two MYO18A genes in zebrafish (Cao et al., 2014, 2016). Using in situ hybridization they detected ubiquitous expression of both genes, with enrichment in the somites. Knockdown of the expression of both of the MYO18A genes using morpholino oligos resulted in disrupted integrity of myofibers, decreased adhesion of myoblasts on laminin substrate, and disruption of the Golgi. In vertebrates, MYO18A apparently has multiple, non-redundant functions.
9.3 MYO18A in Humans
In humans, the ExAC database catalogs mutations found in unbiased whole exome sequencing of a large number of humans (Lek et al., 2016). The ExAC group found that the frequency of inactivating mutations found in MYO18A is exceptionally low, even in persons that have a second, wild-type gene. They conclude with an extremely high degree of statistical confidence that MYO18A exhibits haploinsufficiency in humans. Apparently even a subtle change in MYO18A levels due to loss of a single allele results in lethality, consistent with the idea that MYO18A carries out an essential function.
10. Conclusion
MYO18A is an essential myosin in higher eukaryotes. It plays a role in a variety of intracellular transport processes, including retrograde treadmilling of actin and transport of the PAK2/βPIX/GIT1 complex from focal adhesions to membrane ruffles. A high proportion of MYO18A is found in a complex with GOLPH3, functioning at the trans Golgi to participate in driving vesicle budding for forward trafficking to the plasma membrane. Whether these transport functions of MYO18A involve direct motor function by MYO18A or an interaction with another myosin motor remains uncertain. Nevertheless, it is clear that MYO18A is a unique and interesting protein at the crossroads of signal transduction and intracellular transport.
Acknowledgments
Funding
This work was supported by NIH grants (R01 GM120055 and R01 CA201303), an Era of Hope Scholar Award from the Department of Defense Breast Cancer Research Program (W81XWH-10-1-0822), a Burroughs Wellcome Fund Career Award in the Biomedical Sciences, and a Scholar-Innovator Award from the Harrington Discovery Institute.
We thank members of the Field lab for critical reading of the manuscript.
Footnotes
Conflict of interest
We have no conflicts to report.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Audhya A, Foti M, Emr SD. Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol Biol Cell. 2000;11:2673–2689. doi: 10.1091/mbc.11.8.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AW, Ward MA, Blackstock WP, Freeman HN, Choudhary JS, Lewis AP, Chotai D, Fazel A, Gushue JN, Paiement J, Palcy S, Chevet E, Lafrenière-Roula M, Solari R, Thomas DY, Rowley A, Bergeron JJ. Proteomics characterization of abundant Golgi membrane proteins. J Biol Chem. 2001;276:5152–5165. doi: 10.1074/jbc.M006143200. [DOI] [PubMed] [Google Scholar]
- Billington N, Beach JR, Heissler SM, Remmert K, Guzik-Lendrum S, Nagy A, Takagi Y, Shao L, Li D, Yang Y, Zhang Y, Barzik M, Betzig E, Hammer JA, Sellers JR. Myosin 18A coassembles with nonmuscle myosin 2 to form mixed bipolar filaments. Curr Biol CB. 2015;25:942–948. doi: 10.1016/j.cub.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishé B, Syed GH, Field SJ, Siddiqui A. Role of phosphatidylinositol 4-phosphate (PI4P) and its binding protein GOLPH3 in hepatitis C virus secretion. J Biol Chem. 2012;287:27637–27647. doi: 10.1074/jbc.M112.346569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonn BR, Rudolf A, Hornbruch-Freitag C, Daum G, Kuckwa J, Kastl L, Buttgereit D, Renkawitz-Pohl R. Myosin heavy chain-like localizes at cell contact sites during Drosophila myoblast fusion and interacts in vitro with Rolling pebbles 7. Exp Cell Res. 2013;319:402–416. doi: 10.1016/j.yexcr.2012.12.005. [DOI] [PubMed] [Google Scholar]
- Buschman MD, Rahajeng J, Field SJ. GOLPH3 links the Golgi, DNA damage, and cancer. Cancer Res. 2015;75:624–627. doi: 10.1158/0008-5472.CAN-14-3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Li S, Shao M, Cheng X, Xu Z, Shi D. The PDZ-containing unconventional myosin XVIIIA regulates embryonic muscle integrity in zebrafish. J Genet Genomics Yi Chuan Xue Bao. 2014;41:417–428. doi: 10.1016/j.jgg.2014.06.008. [DOI] [PubMed] [Google Scholar]
- Cao JM, Cheng XN, Li SQ, Heller S, Xu ZG, Shi DL. Identification of novel MYO18A interaction partners required for myoblast adhesion and muscle integrity. Sci Rep. 2016;6:36768. doi: 10.1038/srep36768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross M, Csar XF, Wilson NJ, Manes G, Addona TA, Marks DC, Whitty GA, Ashman K, Hamilton JA. A novel 110 kDa form of myosin XVIIIA (MysPDZ) is tyrosine-phosphorylated after colony-stimulating factor-1 receptor signalling. Biochem J. 2004;380:243–253. doi: 10.1042/BJ20031978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Gräf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, Langerød A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Børresen-Dale A-L, Brenton JD, Tavaré S, Caldas C, Aparicio S METABRIC Group. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dippold HC, Ng MM, Farber-Katz SE, Lee SK, Kerr ML, Peterman MC, Sim R, Wiharto PA, Galbraith KA, Madhavarapu S, Fuchs GJ, Meerloo T, Farquhar MG, Zhou H, Field SJ. GOLPH3 bridges phosphatidylinositol-4-phosphate and actomyosin to stretch and shape the Golgi to promote budding. Cell. 2009;139:337–351. doi: 10.1016/j.cell.2009.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doberstein SK, Fetter RD, Mehta AY, Goodman CS. Genetic analysis of myoblast fusion: blown fuse is required for progression beyond the prefusion complex. J Cell Biol. 1997;136:1249–1261. doi: 10.1083/jcb.136.6.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea G, Lázaro-Diéguez F, Vilella M. Actin dynamics at the Golgi complex in mammalian cells. Curr Opin Cell Biol. 2006;18:168–178. doi: 10.1016/j.ceb.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Eswaran J, Patnaik D, Filippakopoulos P, Wang F, Stein RL, Murray JW, Higgins JMG, Knapp S. Structure and functional characterization of the atypical human kinase haspin. Proc Natl Acad Sci U S A. 2009;106:20198–20203. doi: 10.1073/pnas.0901989106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber-Katz SE, Dippold HC, Buschman MD, Peterman MC, Xing M, Noakes CJ, Tat J, Ng MM, Rahajeng J, Cowan DM, Fuchs GJ, Zhou H, Field SJ. DNA damage triggers Golgi dispersal via DNA-PK and GOLPH3. Cell. 2014;156:413–427. doi: 10.1016/j.cell.2013.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa T, Ikawa S, Yanai N, Obinata M. Isolation of a novel PDZ-containing myosin from hematopoietic supportive bone marrow stromal cell lines. Biochem Biophys Res Commun. 2000;270:67–75. doi: 10.1006/bbrc.2000.2377. [DOI] [PubMed] [Google Scholar]
- Godi A, Di Campli A, Konstantakopoulos A, Di Tullio G, Alessi DR, Kular GS, Daniele T, Marra P, Lucocq JM, De Matteis MA. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat Cell Biol. 2004;6:393–404. doi: 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- Godi A, Pertile P, Meyers R, Marra P, Di Tullio G, Iurisci C, Luini A, Corda D, De Matteis MA. ARF mediates recruitment of PtdIns-4-OH kinase-beta and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat Cell Biol. 1999;1:280–287. doi: 10.1038/12993. [DOI] [PubMed] [Google Scholar]
- Govindan B, Bowser R, Novick P. The role of Myo2, a yeast class V myosin, in vesicular transport. J Cell Biol. 1995;128:1055–1068. doi: 10.1083/jcb.128.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzik-Lendrum S, Heissler SM, Billington N, Takagi Y, Yang Y, Knight PJ, Homsher E, Sellers JR. Mammalian myosin-18A, a highly divergent myosin. J Biol Chem. 2013;288:9532–9548. doi: 10.1074/jbc.M112.441238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzik-Lendrum S, Nagy A, Takagi Y, Houdusse A, Sellers JR. Drosophila melanogaster Myosin-18 Represents a Highly Divergent Motor with Actin Tethering Properties. J Biol Chem. 2011;286:21755–21766. doi: 10.1074/jbc.M111.218669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halberg N, Sengelaub CA, Navrazhina K, Molina H, Uryu K, Tavazoie SF. PITPNC1 Recruits RAB1B to the Golgi Network to Drive Malignant Secretion. Cancer Cell. 2016;29:339–353. doi: 10.1016/j.ccell.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama H, Schnieders EA, Thorner J, Takemoto JY, DeWald DB. Direct involvement of phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J Biol Chem. 1999;274:34294–34300. doi: 10.1074/jbc.274.48.34294. [DOI] [PubMed] [Google Scholar]
- Harris BZ, Lim WA. Mechanism and role of PDZ domains in signaling complex assembly. J Cell Sci. 2001;114:3219–3231. doi: 10.1242/jcs.114.18.3219. [DOI] [PubMed] [Google Scholar]
- Hirschberg K, Miller CM, Ellenberg J, Presley JF, Siggia ED, Phair RD, Lippincott-Schwartz J. Kinetic analysis of secretory protein traffic and characterization of golgi to plasma membrane transport intermediates in living cells. J Cell Biol. 1998;143:1485–1503. doi: 10.1083/jcb.143.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu RM, Hsieh YJ, Yang TH, Chiang YC, Kan CY, Lin YT, Chen JT, Yu JS. Binding of the extreme carboxyl-terminus of PAK-interacting exchange factor β (βPIX) to myosin 18A (MYO18A) is required for epithelial cell migration. Biochim Biophys Acta. 2014;1843:2513–2527. doi: 10.1016/j.bbamcr.2014.06.023. [DOI] [PubMed] [Google Scholar]
- Hsu RM, Tsai MH, Hsieh YJ, Lyu PC, Yu JS. Identification of MYO18A as a novel interacting partner of the PAK2/betaPIX/GIT1 complex and its potential function in modulating epithelial cell migration. Mol Biol Cell. 2010;21:287–301. doi: 10.1091/mbc.E09-03-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaji T, Im S, Gu W, Wang Y, Hang Q, Lu J, Fukuda T, Hashii N, Takakura D, Kawasaki N, Miyoshi H, Gu J. An oncogenic protein Golgi phosphoprotein 3 up-regulates cell migration via sialylation. J Biol Chem. 2014;289:20694–20705. doi: 10.1074/jbc.M113.542688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogawa Y, Kon T, Inoue T, Ohkura R, Yamakawa H, Ohara O, Sutoh K. The N-terminal domain of MYO18A has an ATP-insensitive actin-binding site. Biochemistry (Mosc) 2005;44:6190–6196. doi: 10.1021/bi0475931. [DOI] [PubMed] [Google Scholar]
- Ivanov AI, Rovescalli AC, Pozzi P, Yoo S, Mozer B, Li HP, Yu SH, Higashida H, Guo V, Spencer M, Nirenberg M. Genes required for Drosophila nervous system development identified by RNA interference. Proc Natl Acad Sci U S A. 2004;101:16216–16221. doi: 10.1073/pnas.0407188101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston GC, Prendergast JA, Singer RA. The Saccharomyces cerevisiae MYO2 gene encodes an essential myosin for vectorial transport of vesicles. J Cell Biol. 1991;113:539–551. doi: 10.1083/jcb.113.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupfer A, Louvard D, Singer SJ. Polarization of the Golgi apparatus and the microtubule-organizing center in cultured fibroblasts at the edge of an experimental wound. Proc Natl Acad Sci U S A. 1982;79:2603–2607. doi: 10.1073/pnas.79.8.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lázaro-Diéguez F, Colonna C, Cortegano M, Calvo M, Martínez SE, Egea G. Variable actin dynamics requirement for the exit of different cargo from the trans-Golgi network. FEBS Lett. 2007;581:3875–3881. doi: 10.1016/j.febslet.2007.07.015. [DOI] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lu Y, Zhang J, Kang H, Qin Z, Chen C. PI4KIIalpha is a novel regulator of tumor growth by its action on angiogenesis and HIF-1alpha regulation. Oncogene. 2010;29:2550–2559. doi: 10.1038/onc.2010.14. [DOI] [PubMed] [Google Scholar]
- Lu Z, Joseph D, Bugnard E, Zaal KJ, Ralston E. Golgi complex reorganization during muscle differentiation: visualization in living cells and mechanism. Mol Biol Cell. 2001;12:795–808. doi: 10.1091/mbc.12.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makowska KA, Hughes RE, White KJ, Wells CM, Peckham M. Specific Myosins Control Actin Organization, Cell Morphology, and Migration in Prostate Cancer Cells. Cell Rep. 2015;13:2118–2125. doi: 10.1016/j.celrep.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millarte V, Farhan H. The Golgi in cell migration: regulation by signal transduction and its implications for cancer cell metastasis. ScientificWorldJournal. 2012;2012:498278. doi: 10.1100/2012/498278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Furusawa T, Okubo T, Inoue T, Ikawa S, Yanai N, Mori KJ, Obinata M. Genome structure and differential expression of two isoforms of a novel PDZ-containing myosin (MysPDZ) (Myo18A) J Biochem (Tokyo) 2003;133:405–413. doi: 10.1093/jb/mvg053. [DOI] [PubMed] [Google Scholar]
- Mori K, Matsuda K, Furusawa T, Kawata M, Inoue T, Obinata M. Subcellular localization and dynamics of MysPDZ (Myo18A) in live mammalian cells. Biochem Biophys Res Commun. 2005;326:491–498. doi: 10.1016/j.bbrc.2004.11.058. [DOI] [PubMed] [Google Scholar]
- Mukherjee K, Sharma M, Urlaub H, Bourenkov GP, Jahn R, Südhof TC, Wahl MC. CASK Functions as a Mg2+-independent neurexin kinase. Cell. 2008;133:328–339. doi: 10.1016/j.cell.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng MM, Dippold HC, Buschman MD, Noakes CJ, Field SJ. GOLPH3L antagonizes GOLPH3 to determine Golgi morphology. Mol Biol Cell. 2013;24:796–808. doi: 10.1091/mbc.E12-07-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Png KJ, Halberg N, Yoshida M, Tavazoie SF. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2011;481:190–194. doi: 10.1038/nature10661. [DOI] [PubMed] [Google Scholar]
- Sanchez-Garcia F, Villagrasa P, Matsui J, Kotliar D, Castro V, Akavia UD, Chen BJ, Saucedo-Cuevas L, Rodriguez Barrueco R, Llobet-Navas D, Silva JM, Pe’er D. Integration of genomic data enables selective discovery of breast cancer drivers. Cell. 2014;159:1461–1475. doi: 10.1016/j.cell.2014.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Tirado FH, Legesse-Miller A, Schott D, Bretscher A. PI4P and Rab inputs collaborate in myosin-V-dependent transport of secretory compartments in yeast. Dev Cell. 2011;20:47–59. doi: 10.1016/j.devcel.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott D, Ho J, Pruyne D, Bretscher A. The Cooh-Terminal Domain of Myo2p, a Yeast Myosin V, Has a Direct Role in Secretory Vesicle Targeting. J Cell Biol. 1999;147:791–808. doi: 10.1083/jcb.147.4.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KL, Kabbarah O, Liang MC, Ivanova E, Anagnostou V, Wu J, Dhakal S, Wu M, Chen S, Feinberg T, Huang J, Saci A, Widlund HR, Fisher DE, Xiao Y, Rimm DL, Protopopov A, Wong KK, Chin L. GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer. Nature. 2009;459:1085–1090. doi: 10.1038/nature08109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever-Chroneos Z, Krupa A, Davis J, Hasan M, Yang CH, Szeliga J, Herrmann M, Hussain M, Geisbrecht BV, Kobzik L, Chroneos ZC. Surfactant protein A (SP-A)-mediated clearance of Staphylococcus aureus involves binding of SP-A to the staphylococcal adhesin eap and the macrophage receptors SP-A receptor 210 and scavenger receptor class A. J Biol Chem. 2011;286:4854–4870. doi: 10.1074/jbc.M110.125567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng G, Zeng Z, Pan J, Kou L, Wang Q, Yao H, Wen L, Ma L, Wu D, Qiu H, Chen S. Multiple MYO18A-PDGFRB fusion transcripts in a myeloproliferative neoplasm patient with t(5;17)(q32;q11) Mol Cytogenet. 2017;10:4. doi: 10.1186/s13039-017-0306-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F, Telesco SE, Liu Y, Radhakrishnan R, Lemmon MA. ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proc Natl Acad Sci U S A. 2010;107:7692–7697. doi: 10.1073/pnas.1002753107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft MH, Behrmann E, Munske-Weidemann LC, Thiel C, Raunser S, Manstein DJ. Functional characterization of human myosin-18A and its interaction with F-actin and GOLPH3. J Biol Chem. 2013;288:30029–30041. doi: 10.1074/jbc.M113.497180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan I, Yong J, Dong JM, Lim L, Leung T. A tripartite complex containing MRCK modulates lamellar actomyosin retrograde flow. Cell. 2008;135:123–136. doi: 10.1016/j.cell.2008.09.018. [DOI] [PubMed] [Google Scholar]
- Tokuda E, Itoh T, Hasegawa J, Ijuin T, Takeuchi Y, Irino Y, Fukumoto M, Takenawa T. Phosphatidylinositol 4-phosphate in the Golgi apparatus regulates cell-cell adhesion and invasive cell migration in human breast cancer. Cancer Res. 2014;74:3054–3066. doi: 10.1158/0008-5472.CAN-13-2441. [DOI] [PubMed] [Google Scholar]
- Ussowicz M, Jaśkowiec A, Meyer C, Marschalek R, Chybicka A, Szczepański T, Haus O. A three-way translocation of MLL, MLLT11, and the novel reciprocal partner gene MYO18A in a child with acute myeloid leukemia. Cancer Genet. 2012;205:261–265. doi: 10.1016/j.cancergen.2012.02.006. [DOI] [PubMed] [Google Scholar]
- Walch-Solimena C, Novick P. The yeast phosphatidylinositol-4-OH kinase pik1 regulates secretion at the Golgi. Nat Cell Biol. 1999;1:523–525. doi: 10.1038/70319. [DOI] [PubMed] [Google Scholar]
- Walz C, Chase A, Schoch C, Weisser A, Schlegel F, Hochhaus A, Fuchs R, Schmitt-Gräff A, Hehlmann R, Cross NCP, Reiter A. The t(8;17)(p11;q23) in the 8p11 myeloproliferative syndrome fuses MYO18A to FGFR1. Leukemia. 2005;19:1005–1009. doi: 10.1038/sj.leu.2403712. [DOI] [PubMed] [Google Scholar]
- Walz C, Haferlach C, Hänel A, Metzgeroth G, Erben P, Gosenca D, Hochhaus A, Cross NCP, Reiter A. Identification of a MYO18A-PDGFRB fusion gene in an eosinophilia-associated atypical myeloproliferative neoplasm with a t(5;17)(q33-34;q11.2) Genes Chromosomes Cancer. 2009;48:179–183. doi: 10.1002/gcc.20629. [DOI] [PubMed] [Google Scholar]
- Wu CC, Taylor RS, Lane DR, Ladinsky MS, Weisz JA, Howell KE. GMx33: a novel family of trans-Golgi proteins identified by proteomics. Traffic Cph Den. 2000;1:963–975. [PubMed] [Google Scholar]
- Xing M, Peterman MC, Davis RL, Oegema K, Shiau AK, Field SJ. GOLPH3 drives cell migration by promoting Golgi reorientation and directional trafficking to the leading edge. Mol Biol Cell. 2016;27:3828–3840. doi: 10.1091/mbc.E16-01-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, English JM, Wilsbacher JL, Stippec S, Goldsmith EJ, Cobb MH. WNK1, a novel mammalian serine/threonine protein kinase lacking the catalytic lysine in subdomain II. J Biol Chem. 2000;275:16795–16801. doi: 10.1074/jbc.275.22.16795. [DOI] [PubMed] [Google Scholar]
- Yang CH, Szeliga J, Jordan J, Faske S, Sever-Chroneos Z, Dorsett B, Christian RE, Settlage RE, Shabanowitz J, Hunt DF, Whitsett JA, Chroneos ZC. Identification of the surfactant protein A receptor 210 as the unconventional myosin 18A. J Biol Chem. 2005;280:34447–34457. doi: 10.1074/jbc.M505229200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Carrillo M, Wu YM, DiAngelo SL, Silveyra P, Umstead TM, Halstead ES, Davies ML, Hu S, Floros J, McCormack FX, Christensen ND, Chroneos ZC. SP-R210 (Myo18A) Isoforms as Intrinsic Modulators of Macrophage Priming and Activation. PloS One. 2015;10:e0126576. doi: 10.1371/journal.pone.0126576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P. The prevalence and significance of PDZ domain-phosphoinositide interactions. Biochim Biophys Acta. 2006;1761:947–956. doi: 10.1016/j.bbalip.2006.04.003. [DOI] [PubMed] [Google Scholar]