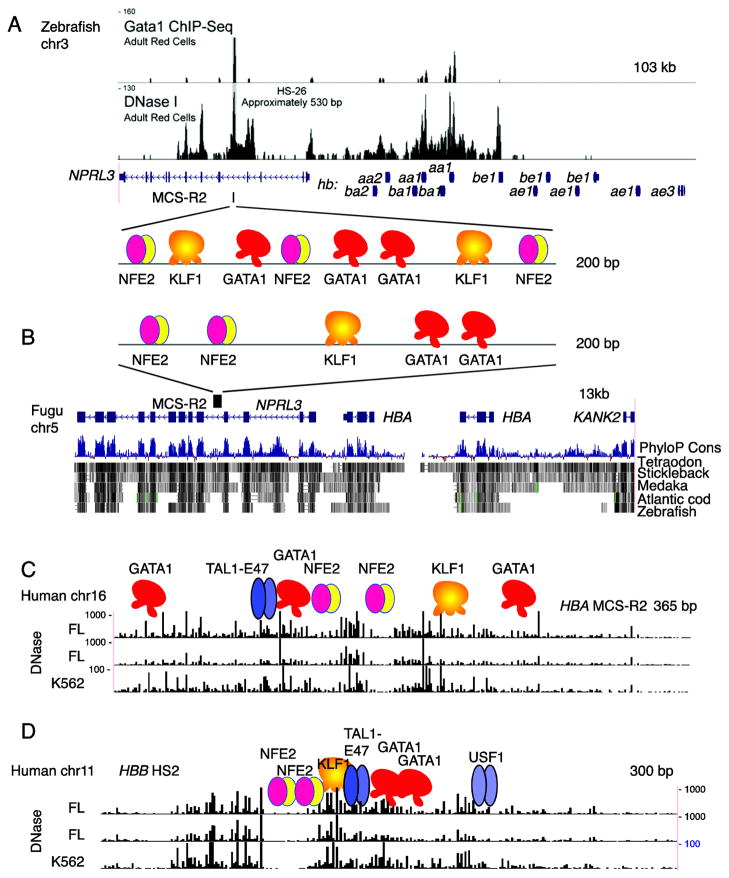
Figure 4. Conservation of epigenetic signals versus genome sequences in distal regulatory regions of globin gene clusters.
(A) Binding of GATA1 and DNase accessibility at the major regulatory element (MCS-R2) for the globin gene cluster on chromosome 3 of zebrafish. The signal tracks are from Figure 4 of Ganis et al. [10]; they are aligned with a gene map from the genome assembly. The proteins inferred to be bound at MCS-R2 are shown as colored icons in the zoomed-in view. (B) Genome sequence conservation and divergence at MCS-R2 in other fish. The HBA cluster from Fugu on chromosome 5 is shown, highlighting the MCS-R2 by showing the inferred proteins bound. Underneath the gene map is a track showing the likelihood that DNA segments are under purifying selection. That PhyloP Cons score is estimated from sequence alignments of multiple species, which are shown as dark rectangles indicating aligned sequences. Note that the intron containing the Fugu MCS-R2 aligns with sequences from three other fish, but not to zebrafish. (C) DNase footprints for the HBA MCS-R2 in humans. The high density DNase-seq analysis [63, 64] was done on highly erythroid human fetal liver (FL) tissues and in K562 cells. Regions of frequent cleavage (greater accessibility in chromatin) have a high signal on the tracks. Positions of bound transcription factors, determined from other studies, are shown as colored icons. (D) DNase footprints for the HBB LCR 5′HS2 in humans.