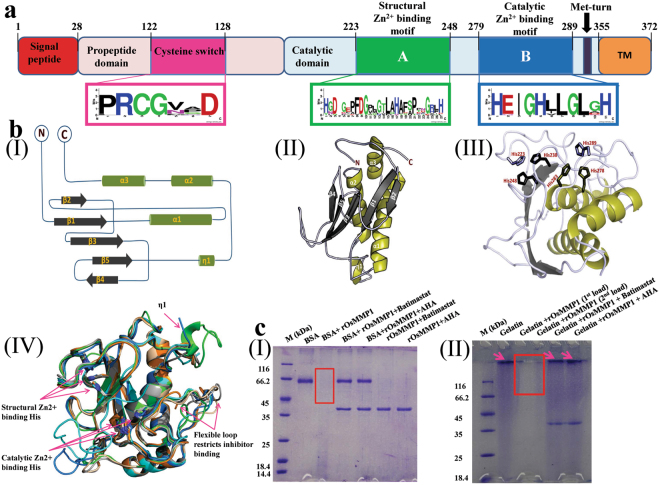
Figure 1.
Structure prediction and proteolytic activity assay of OsMMP1. (a) Schematic diagram of the predicted domains of OsMMP1 protein and WebLogo plot of the consensus sequence of cysteine switch, structural and catalytic Zn2+-binding motifs. The consensus sequence was determined based on the frequency of each amino acid in corresponding position of the amino acid sequence of the aligned MMPs using WebLogo design tool (http://weblogo.berkeley.edu/logo.cgi). WebLogo plot reveals that the niches having the cysteine switch (PRCGVAD) and catalytic Zn2+-binding motif (HEIGHLLGLGH) are highly conserved in comparison with the structural Zn2+-binding motif (HGDGEAFDGPLGTLAHAFSPTDGRFH). The diagram is not drawn to the scale. The number indicates the position of amino acids spanning the critical domains and motifs. (bI) Topology diagram of the OsMMP1 catalytic domain displays four parallel β-sheets, one anti-parallel β sheet, three α- helices and a 310-helix (η1). (bII) Cartoon representation of the model structure of OsMMP1 catalytic domain. (bIII) The 3D orientation of six His residues participating in the coordination bond with two Zn2+ ions. (bIV) Structural superimposition of OsMMP1 (green) with human MMP1 (red), MMP2 (marine), MMP3 (wheat), MMP9 (cyan), MMP10 (orange), and MMP13 (grey) shows the conserved folds and the conserved secondary structures. (cI,II) Analysis of the products formed after protease activity of the recombinant OsMMP1 (rOsMMP1). Rectangular boxes indicate the proteolytic degradation of (cI) BSA and (cII) gelatin. The arrow indicates the (cII) gelatin protein band. Lane M: protein molecular weight marker. The degradation of (cI) BSA is prominent in the 3rd lane, but the rOsMMP1 band is absent due to its autocatalytic property. Similarly, the degradation of (cII) gelatin is prominent in the 3rd and 4th lanes, but the rOsMMP1 band is absent due to its autocatalytic property. The MMP inhibitors, Batimastat and acetohydroxamic acid (AHA) are efficient in inhibiting the proteolytic and autocatalytic activities of rOsMMP1. Effects of both the inhibitors are quite similar as both of them completely inhibit the activity of rOsMMP1 but the concentration of AHA is 25 times higher than Batimastat. Full-length gels of cI and cII are presented in Supplementary Figs S16, and S17, respectively.