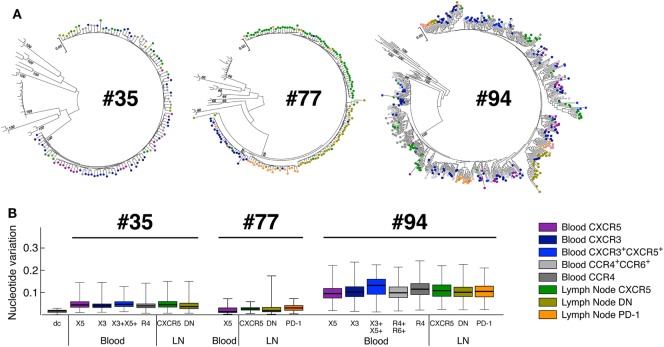
Figure 5.
Phylogenetic relationship of HIV-1 envelope sequences derived from CD4 T-cell populations. Sequences of the highly variable EnvV1V4 region (Hxb 6540-7668) were derived from blood and LN memory CD4 T-cell populations isolated from aviremic-treated HIV-1-infected individuals (N = 3). Virus quasi-species were amplified and sequenced [single molecule, real-time (SMRT) method/PacBio Systems]. (A) Phylogenetic relationship of HIV-1 envelope sequences derived from CD4 T-cell populations (N = 3). The phylogenetic relationship was inferred by the Maximum Likelihood method based on the General Time Reversible substitution model (GTR + G). Each tree includes reference sequences from subtype B and non-B HIV-1-infected individuals. (B) Nucleotide variations observed within each blood and LN memory CD4 T-cell population estimated using the Gama distributed kimura-two-parameter. CD4 T-cell populations were color coded (A,B). dc corresponds to the laboratory isolate control used to monitor sequence diversity induced by the method. “X3” corresponds to blood CXCR3-expressing CD4 T cells; “R4” corresponds to blood CCR4-expressing CD4 T cells; R4+R6+ corresponds to blood CCR4+CCR6+ CD4 T cells; “X5” corresponds to blood CXCR5-expressing CD4 T cells; And X3+X5+ corresponds to blood CXCR3+CXCR5+ CD4 T cells. “LN” corresponds to lymph node.