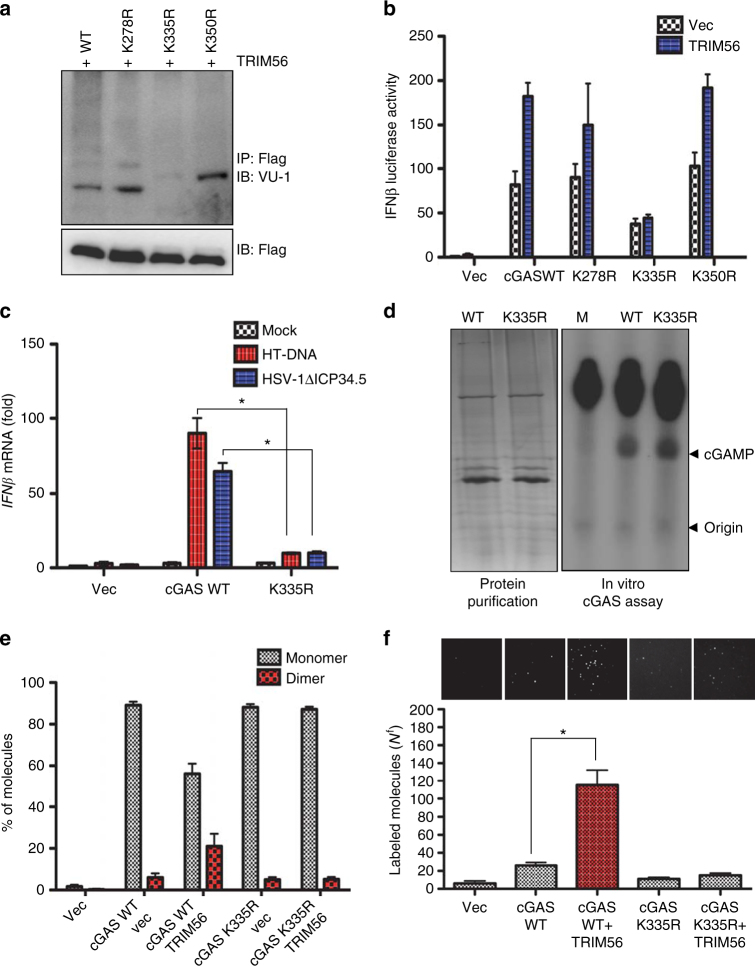
Fig. 4.
TRIM56 increases cGAS dimerization and DNA-binding ability. a HEK293T cells were transfected with cGAS WT or cGAS K→R mutants. WCLs were used for immunoprecipitation and immunoblotting, as indicated. b IFNβ promoter activity in 293T cells, which were co-transfected with TRIM56 and cGAS WT or K→R mutants. c L929 cGAS−/− cells were stably complemented with empty vector, mouse cGAS WT, or cGAS K335R. Cells were stimulated with HT-DNA or infected with HSV-1ΔICP34.5 (MOI = 5). IFNβ mRNA was analyzed by RT-PCR. d In vitro enzymatic assays were performed in the presence of P32-α-GTP with amino acids 141–507 of mouse cGAS purified from engineered E. coli BL21 strain. The left panel shows the purified cGAS WT and cGAS K335R proteins. cGAMP production was analyzed by TLC and autoradiography. The bottom arrow shows the spotted origin and the top arrow shows the migrated cGAMP. e The relative ratio of observed bleaching steps for monomeric cGAS-GFP and dimeric cGAS-GFP pulled down from cell lysates. HEK293T cells were transfected with vector, cGAS WT, cGAS WT/TRIM56, cGASK335R, or cGAS335R/TRIM56, and the ratio was calculated by counting the photobleaching steps from a GFP-cGAS pull-down. For determining the stoichiometry, traces were manually scored for the number of bleaching steps. More than 300 traces were scored to reliably identify the photobleaching step distribution. See Supplementary Fig. 6. f Rhodamine-labeled double-stranded DNA bound by cGAS on the PEG-coated surface. Top panel: representative fluorescent DNA images bound by cGAS pulled down from cell lysates from vector; cGAS, cGAS- and TRIM56-expressing cells; cGASK335R-; or cGASK335R- and TRIM56-expressing cells. Rhodamine-labeled pdA:dT (1 μg/ml) was co-transfected into vector, cGAS, TRIM56/cGAS-expressing cells, cGASK335R, or cGASK335R/TRIM56-expressing cells, respectively. Bottom panel: average number of molecules per imaging area (Nf). Error bars indicate mean ± s.d. *P < 0.05, Student’s t-test. Data in a, d are representative of two independent experiments. Data in b, c, e, f are representative of three independent experiments. Error bars b, c, e, f indicate mean ± s.d. of n = 3. *P < 0.05, versus control using Student’s t-test (c, f). Full blots are shown in Supplementary Fig. 10