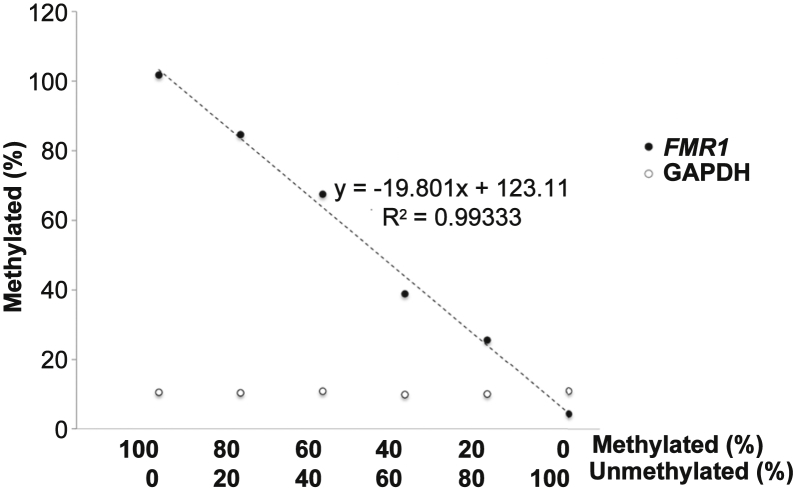
Figure 3.
Analysis of different ratios of methylated and unmethylated alleles using the qMS-PCR assay. DNA from a fully methylated full mutation carrier (GM09145) and a full unmethylated normal allele (GM06865) was mixed at different ratios before sonication. The DNA mixtures were then either mock digested or digested with HpaII and the amount of DNA corresponding to the 5′ end of the FMR1 gene analyzed by real-time PCR, as described in Materials and Methods. The percentage of uncut FMR1 DNA (ie, the percentage methylated) was then plotted as a function of the relative amount of the original methylated and unmethylated DNA (closed circles). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH; open circles) was analyzed in parallel as a control for HpaII digestion.