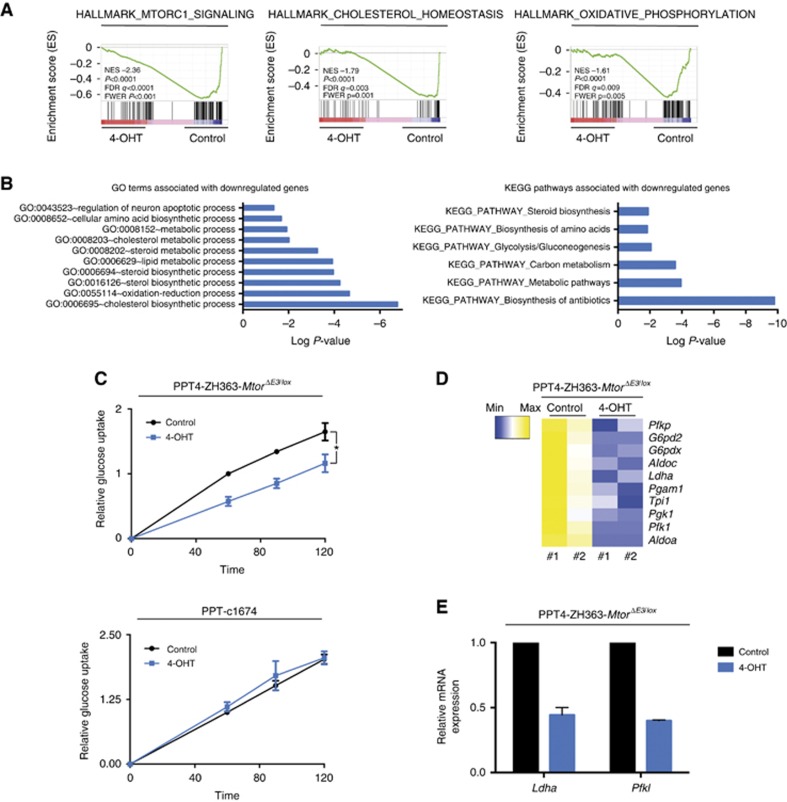
Figure 3.
Mtor is connected to metabolic pathways. (A) PPT4-ZH363-MtorΔE3/lox cells were treated with vehicle or 4-OHT (600 nM) over 8 days. Afterwards, mRNA expression of two biological replicates was profiled using RNA-Seq and analysed by GSEA. The normalised enrichment score (NES), nominal P-value, FDR q-value and FWER P-values are indicated. (B) GO-Term and KEGG pathway analysis was conducted with genes downregulated (log 2FC⩽−0.58) upon the Mtor deletion using the RNA-Seq data from (A). Terms and pathways with a Benjamini-corrected P-value <0.05 were depicted. (C) PPT4-ZH363-MtorΔE3/lox or control PPT-c1674 cells were treated with vehicle or 4-OHT (600 nM) over 8 days. Afterwards, equal number of cells was analysed for glucose uptake over time using F-18-FDG uptake assay. *P-value of an analysis of variance (ANOVA) test <0.05 (n=4). (D) Heat map of glycolytic enzymes from RNA-Seq data described in (A). (E) PPT4-ZH363-MtorΔE3/lox cells were treated with vehicle or 4-OHT (600 nM) over 8 days. Afterwards, mRNA expression of Ldha and PfkI mRNA expression was determined by qPCR using beta-actin mRNA expression as reference (n=2).