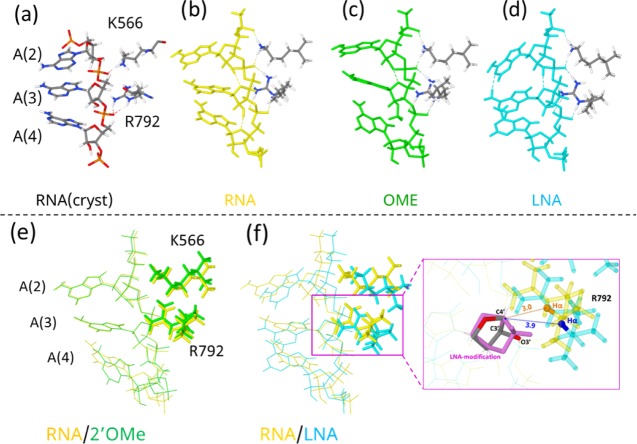
Figure 5.
Computational prediction of the unmodified, 2′-OMe-modified, and LNA-modified structures loaded on the AGO protein binding pocket. Unmodified AAA RNA with the R792 and K566 crystal structures from Schirle et al.12 (a), optimized structure of unmodified (b), 2′-OMe-modified (c), and LNA-modified RNA and (d) RNA with R792 and K566. Superposition at C4′-C3′-O3′ of A(3) of unmodified RNA with 2′-OMe-modified RNA (e) and unmodified RNA with LNA-modified RNA (f). The numbers in (f) are the distances (in Å) between C4′ (of A(3)) and Hα (of R792) atoms in unmodified RNA (in brown) and in LNA-modified RNA (in blue).