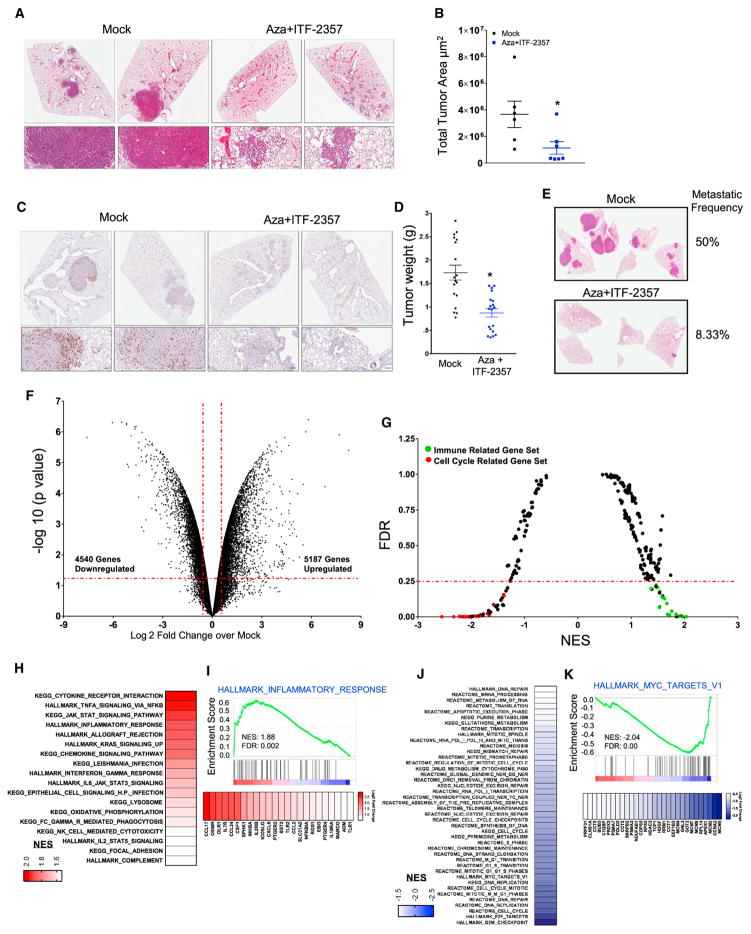
Figure 5. Combination Epigenetic Treatment Reduces Lung Tumor Burden and Progression in Mouse Models of NSCLC.
(A) Representative H&E-stained images of lung sections from mice treated with mock or Aza + ITF-2357. Scale bar, 100 μm.
(B) Quantitation of total tumor area occupied by lesions in lungs of LSL-KrasG12D mice treated with mock or Aza + ITF-2357. Data are presented as mean ± SEM (p value determined by two-tailed t test; n = 6 mock and n = 7 Aza + ITF-2357 mice per group/2 sections analyzed per mouse).
(C) Representative Ki67-stained IHC images of lung sections from mice treated with mock or Aza + ITF-2357 (n = 5 per group). Scale bar, 100 μm.
(D) Lewis Lung Carcinoma (LLC) tumor weights of subcutaneous explants from 1-month mock- and Aza + ITF-2357-treated mice (n = 19 mice per group; error bars, SEM).
(E) Representative H&E-stained images of lung metastasis from LLC mice obtained from 1-month mock- and Aza + ITF-2357-treated mice (n = 12 mice per group), with the indicated percentage frequency of metastasis.
(F) Volcano plot of relative RNA expression from LSL-KrasG12D mouse lung tumors treated for 3 months with Aza + ITF-2357 as compared to mock mice. Genes in upper left and right quadrants are significantly differentially expressed (microarray, n = 2 per group).
(G) GSEA (KEGG, REACTOME, and HALLMARK) pathway distribution for Aza + ITF-2357 versus mock tumors from LSL-KrasG12D mice. Horizontal line denotes FDR significance cutoff of 0.25. Immune- and cell cycle-related gene sets are demarcated by green and red dots, respectively (microarray, n = 2 per group).
(H) Gene sets upregulated in LSL-KrasG12D mice (FDR < 0.25 and NES > 1.5) by Aza + ITF-2357. Color gradation is based on GSEA NES.
(I) Representative upregulated GSEA plots with corresponding core-enriched genes. Color gradation is representative of Log2 fold change over mock.
(J) Gene sets downregulated in LSL-KrasG12D mice (FDR < 0.25 and NES < 1.5) by Aza + ITF-2357. Color gradation is based on GSEA NES.
(K) Representative downregulated GSEA plot with core-enriched genes. Color gradation is representative of Log2 fold change over mock-treated RNA expression.
*p < 0.05 calculated using two tailed t test. See also Figure S6.