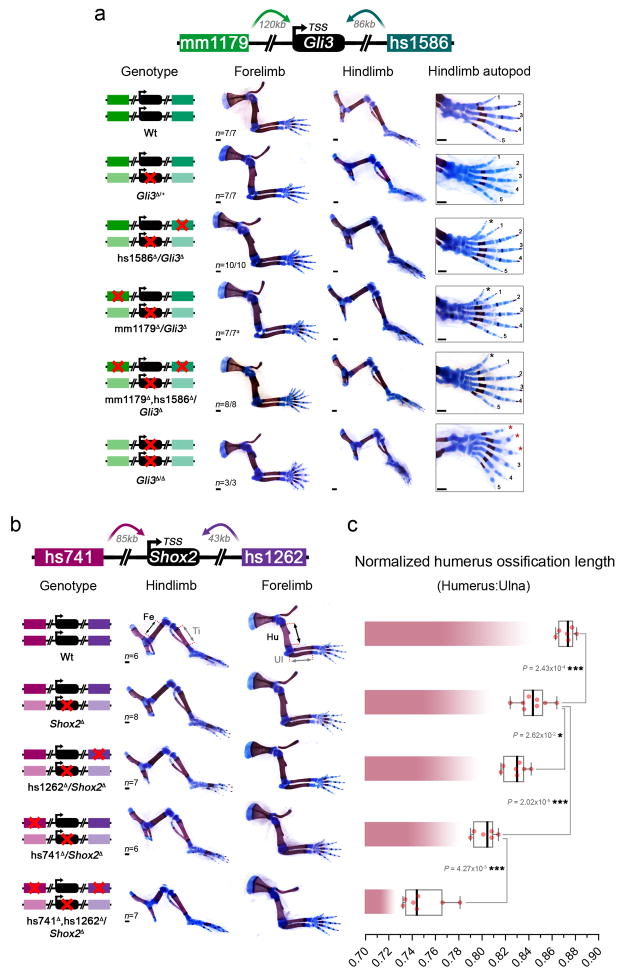
Extended Data Figure 8. Limb phenotypes of individual and combinatorial Gli3 and Shox2 enhancer knockouts in presence of reduced target gene dosage.
(a) Skeletal phenotypes resulting from mm1179 and hs1586 enhancer deletions in combination with reduction to one copy of the Gli3 gene at E18.5. Genotypes are shown on the left with red crosses indicating elements deleted by CRISPR/Cas9. While forelimbs of Gli3Δ/+ embryos displayed bifurcated digit 1 terminal phalanges65, hindlimbs showed an extra toe structure but without detectable cartilage template. Four out of seven mm1179Δ/Gli3Δ embryos displayed additional bifurcation of digit 2 of the right forelimb, which suggests that removal of mm1179 reduces Gli3 levels in the anterior forelimb more than deletion of hs1586. An almost complete anterior extra toe forms in hindlimbs of embryos with single or dual enhancer deletions in the sensitized background (black asterisks). Loss of both Gli3 copies results in anterior hindlimb polydactyly with altered digit identities (red asterisks)24. (b) Allelic series depicts shortening of the stylopod (humerus and femur) in limb skeletons with individual or combined hs741 and hs1262 enhancer deletions in a Shox2 sensitized condition (see also Fig. 3b). Stylopod ossification length (double arrows) appears less reduced in forelimbs (humerus, Hu) than in hindlimbs (femur, Fe) of embryos lacking the activity of both enhancers (hs741Δ, hs1262Δ/Shox2Δ). Tibia (Ti) and ulna (Ul) were normal in all genotypes examined. (c) Humerus ossification length (normalized to ulna ossification length) is significantly reduced in embryos lacking either hs741 or hs1262 in the presence of only one copy of Shox2. In embryos lacking both enhancers in the sensitized background significant shortening of the humerus ossification is observed (compared to all other genotypes). “n” indicates the number of independent biological replicates with similar results. Box plots indicate median, interquartile values, range and individual biological replicates. ***, P < 0.001; *, P < 0.05 (two-tailed, unpaired t-test). Scale bars, 500 μm.