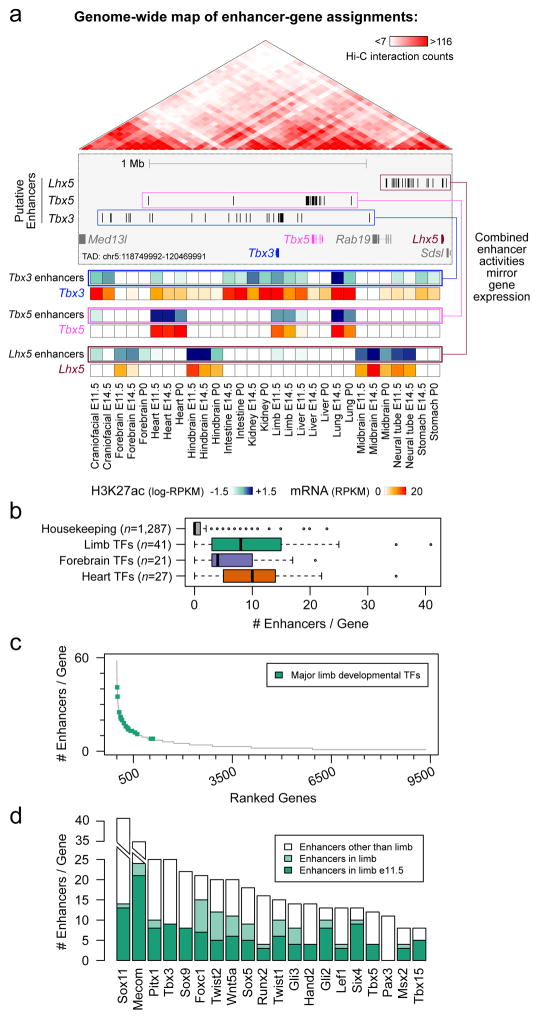
Figure 4. Enhancers with redundant signatures are prevalent near developmental genes.
(a) Enhancer-gene assignments based on correlation of H3K27ac and mRNA profiles across a wide array of tissues (Extended Data Fig. 9a). Top: At an example locus encompassing Tbx3, Tbx5, and Lhx5, up to 25 enhancers are assigned to each of these three genes (blue, pink and brown boxes, Extended Data Fig. 9c). Genes showing fewer than five assigned enhancers are shown in gray. Bottom: heat maps showing meta-profiles of each gene’s expression profile across tissues (red shades), along with the cumulative activity profile of its assigned enhancers (blue shades). (b) Distribution of the number of enhancers assigned to developmental TFs with biased expression in limb (P = 5e-19 vs. housekeeping), forebrain (P = 8e-15), and heart (P = 3e-25) (two sided Mann-Whitney tests). Box plots show median, interquartile values, range, and outliers (individual points). (c) Complete spectrum of genes with at least one assigned enhancer, sorted by decreasing enhancer numbers. Limb-biased TFs are highlighted in green. (d) Total number of enhancers (in all tissues analyzed) assigned to each TF in (c), with the number of assigned enhancers predicted specifically in limb at E11.5 (dark green) or any other stage analyzed (light green).