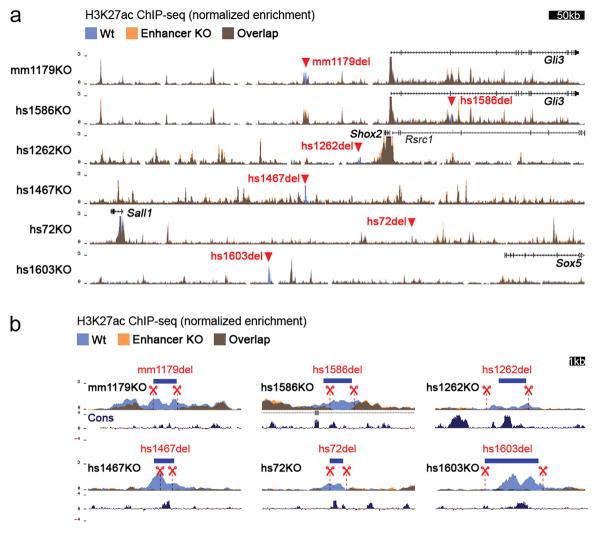
Extended Data Figure 4. Absence of compensatory enhancer signatures in limbs of enhancer KO embryos.
(a) Layered ChIP-seq H3K27 acetylation (ac) profiles surrounding the deleted enhancers and from wild-type (blue, n=4 independent biological replicates) and enhancer knockout embryos (orange, at least n=2 biological replicates). For all samples, E11.5 forelimb was profiled. For display, replicates were merged using bigWigMerge (UCSC tools) and normalized. Red triangles indicate the position of individual enhancer deletions. (b) H3K27ac enrichments in targeted regions marked by red triangles in A, showing the absence of H3K27ac at the deletion site in individual enhancer knockout (orange) compared to wild-type (blue) samples. Blue bars indicate location of enhancer sequences. Dashed red lines demarcate the region deleted by CRISPR. Vertebrate basewise conservation track by PhyloP (Cons) is shown.