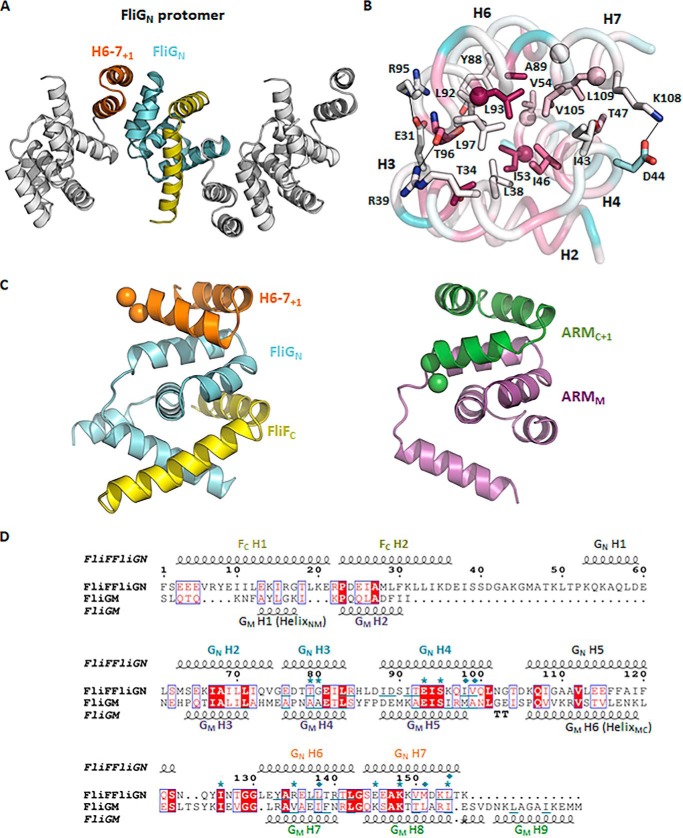
Figure 6.
The ARM-like motif of FliGN associated with helices 6–7, forming a functional unit that shared conserved structural topology with the ARM-like motif of the middle and C-terminal domains. A, a linear array of the FliGN–FliFC complexes in the crystal lattice. Adjacent complex molecules were arranged in opposite orientations. Helices 6–7 are colored in orange; the symmetry molecules are colored in white. B, the binding interface between helices 2–4 and helices 6–7+1 of FliGN. The residues mediating the hydrophobic or electrostatic interactions are shown as sticks. The corresponding residues that were most effectively cross-linked in E. coli (11) are shown as spheres. The structure is colored according to the sequence conservation score calculated by the ConSurf server (26). The sequence alignment was generated by randomly choosing 150 amino acid sequences from the server with sequence identity to FliG between 0.25 and 0.5. C and D, the structural alignment between the FliGN unit and ARMM–ARMC unit. The structural alignment was performed using the PROMALS3D web server (27), and the aligned sequence is represented by ESPript (http://espript.ibcp.fr) (28). (Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.) The conserved diglycine motif is shown as a sphere. The secondary structure of FliFC–FliGN and FliGM is drawn above and below the alignment, respectively. Identical amino acids are in red boxes. The conserved residues are highlighted in red. The residues involved in ARM-like packing are underlined. The FliG mutations that suppressed the clockwise rotational bias of E. coli expressing the FliF–FliG fusion protein are marked with asterisks. The cysteine mutation sites that were most effectively cross-linked are marked with a rhombus (11).