Figure 2. Endothelial cell sprount clustering is partially rescued with αv blocking.
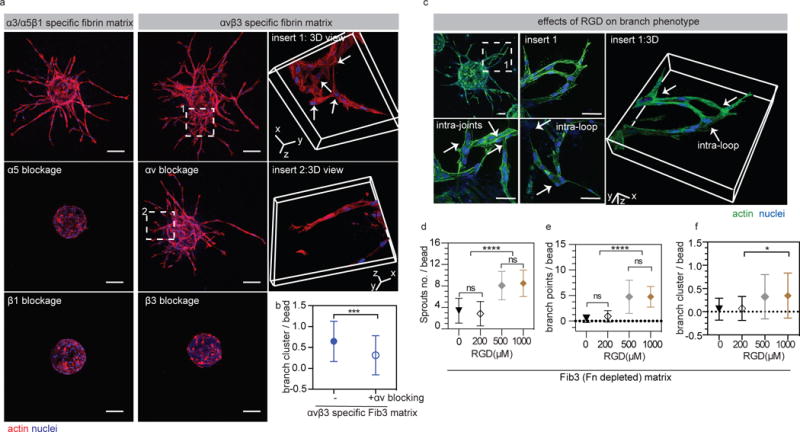
(a) HUVEC sprouting in the presence of integrin αv, α5, β1 or β3 (5μg/ml changed daily) blocking antibodies. Blocking α5, β1 or β3 completely blocks sprouting, while blocking αv does not. α3/α5β1 specific matrices = gel + 2μM Fn9*10, αvβ3 specific matrices = gel + 2μM Fn9(4G)10. Scale bars, 100 μm. (b) comparison of branch cluster occurrence with and without αv blocking (n=51 HUVEC coated beads from three independent gels). (c) Images showing HUVEC sprouts in RGD (1000μM) modified FXIIIa stabilized Fn depleted fibrin matrices. RGD modified matrices promote HUVEC sprouting and branch cluster formation. Scale bar: 50 μm (d,e) Quantification of HUVEC sprouting in RGD modified Fib 3 matrices at different concentrations. (f) Quantification of branch clusters per bead. For quantification n ≥ 15 HUVEC coated beads, from 3 independent gels. Each bead is treated as its own independent sample. Statistical analyses were performed using Prism (GraphPad, San Diego, CA). Brown-Forsythe test was performed to ensure that the variance of the data is similar between the groups that are being statistically compared. Data were analyzed using a one-way analysis of variance (ANOVA) followed by a Tukey post-hoc test and a 95% confidence interval. All plots represent mean ± SD. *,*** and **** indicate P < 0.05, P < 0.001 and P < 0.0001.
