Figure 3. FOXF1 modulates enhancer landscape and ETV1 binding to GIST-lineage specific enhancers.
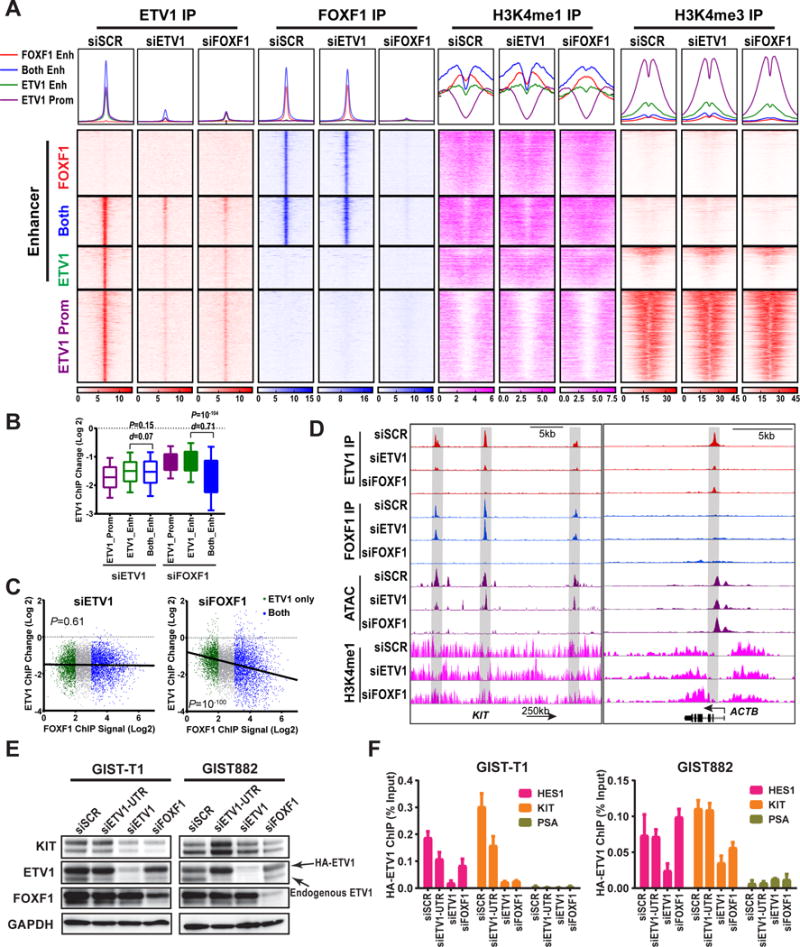
A, ChIP-seq profile (top) and heatmap (bottom) of ETV1, FOXF1, H3K4me1 and H3K4me3 signal around peak center at enhancers (FOXF1 only: red, FOXF1/ETV1-shared [both]: blue and ETV1-only: green) and at ETV1-bound promoters (purple) with siETV1, siFOXF1, or scramble (siSCR) controls in GIST48 cells. B, Box and whisker plots showing change of ETV1-ChIP-seq (Log 2) by siRNA-mediated downregulation of ETV1 or FOXF1 in GIST48 cells. Box 75%, whiskers 90%. P: Mann-Whitney test d: Cohen size-effect. C, Scatter plots of ETV1-ChIP-seq (Log 2) correlated with FOXF1 ChIP-seq signal (Log 2) by siRNA-mediated downregulation of ETV1 or FOXF1 in GIST48 cells. The ETV1-ChIP-signal change is marked green for ETV1-only, and blue for FOXF1and ETV1-shared (both) enhancer peaks. P: Fisher exact test. D, Representative FOXF1, ETV1 and H3K4me1 ChIP-seq and ATAC-seq profiles at the indicated gene loci with siRNA-mediated perturbation of ETV1 and FOXF1 in GIST48 cells. Gray highlighted the enhancers with appreciable changes in ATAC-seq, and H3K4me1 ChIP-seq signals at the FOXF1-regulated KIT locus, but not in the ETV1-regulated and FOXF1-independent HES1 locus. E, Immunoblots of GIST-T1 and GIST882 cells with exogenous expression of Flag-HA-tagged-ETV1 (HA-ETV1) independent of endogenous FOXF1-mediated transcriptional control under experimental perturbations as indicated. F, ChIP-qRT-PCR signals of HA-ETV1 (α-HA ChIP) at the HES1 enhancer (an ETV1-, but not FOXF1-regulated gene), KIT enhancer (a FOXF1- regulated gene) and PSA control (neither ETV1- nor FOXF1-regulated gene) loci under different conditions as indicated in GIST-T1 and GIST882 cells with exogenous expression of HA-ETV1 as in E. N=3, Mean ± SEM.
