Fig. 5. miR-1343-containing exosomes reduce TGF-β signaling and markers of fibrosis.
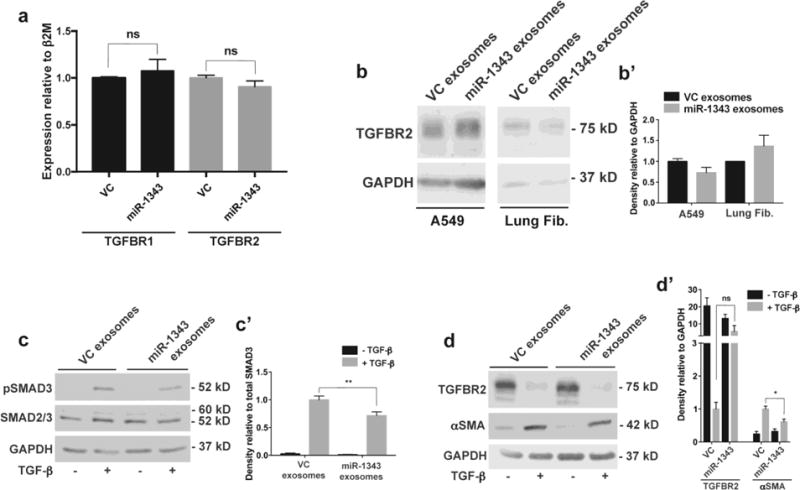
a. TGFBR1 (black bars) and TGFBR2 (grey bars) mRNA measured by qRT-PCR in A549 cells treated for 48 hours with exosomes purified from pCMV-MIR (VC) or pCMV-MIR-1343 (miR-1343) electroporated HL-60 cells. Ct values were normalized to beta-2-microglobulin (β2M). n=4. ns=not significant by unpaired Student’s t test. b. Western blot of lysates from A549 cells (left) and primary lung fibroblasts (right) treated with VC or miR-1343 exosomes as in (a). Blots were probed with antibodies specific for TGFBR2 or GAPDH as the loading control. b’. Proteins were quantified relative to GAPDH and are expressed as fold change compared to VC, n=2. c-c’. Western blot of lysates from primary lung fibroblasts treated with VC or miR-1343 exosomes as in (a) and treated with TGF-β1 (5 ng/mL, +; grey bars) or vehicle control (−; black bars) for 1 hr. (c) Blots were probed with antibodies specific for pSMAD3 (phosphorylated [active] SMAD3), total SMAD2/3 (SMAD2 larger form and SMAD3 below) or GAPDH. (c’) Proteins were quantified relative to total SMAD3 and shown as fold change compared to VC +TGF-β, n=4. **p≤0.01 by ANOVA and Tukey-Kramer post hoc test between TGF-β treatment groups. d-d’. Blots as in (cc’) treated with TGF-β1 for 48 hrs, and (d) probed with antibodies specific for TGFBR2, αSMA, or GAPDH. (d’) Proteins were quantified relative to GAPDH and shown as fold change compared to VC +TGF-β. n=4. *p≤0.05, ns=not significant by ANOVA and Tukey-Kramer post hoc test between TGF-β treatment groups.
