Figure 6.
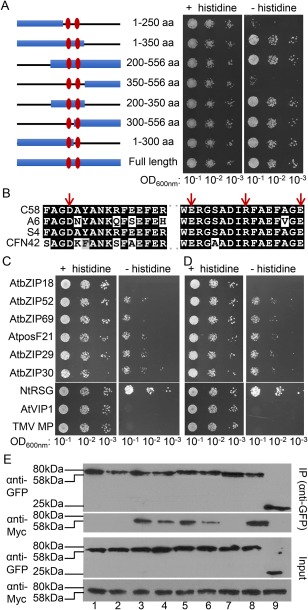
Interaction of C58 VirE2 mutants with AtVIP1 and its homologues. (A) A schematic summary of C58 VirE2 deletion mutants (left) and yeast two‐hybrid assay for interaction between the indicated C58 VirE2 mutants fused to LexA and AtVIP1 fused to Gal4‐AD (right). Numbers represent the positions of amino acid (aa) residues present in each mutant, and the two central domains are indicated with vertical ovals. The indicated dilutions of cell cultures were plated and grown on non‐selective (+histidine) and selective (–histidine) media in the presence of 0.1 mm 3‐amino‐1,2,4‐triazole (3‐AT). (B) Amino acid sequence alignment of the two central domains in C58 VirE2, A6 VirE2, S4 VirE2 and CFN42 VirE2. The residues mutated in the C58 VirE2 4M mutant, i.e. D281, E324, R331 and E338, are indicated with arrows. (C) Yeast two‐hybrid assay for C58 VirE2 4M interactions with AtVIP1 and its homologues. (D) Yeast two‐hybrid assay for C58 VirE2 350–556 interactions with AtVIP1 and its homologues. LexA‐C58 VirE2 4M or LexA‐C58 VirE2 350–556 was co‐expressed with Gal4‐AD fused to the indicated tested proteins. The indicated dilutions of cell cultures were plated and grown on non‐selective (+histidine) and selective (–histidine) media in the presence of 0.1 mm 3‐AT. (E) Co‐immunoprecipitation assay for C58 VirE2 4M interactions with AtVIP1 and its homologues. The experiment was performed using C58 VirE2 4M‐Myc, exactly as described in Fig. 2B for a similar assay with C58 VirE2‐Myc. Top panel: proteins immunoprecipitated (IP) with anti‐GFP antibody and analysed by western blotting with anti‐GFP antibody or anti‐Myc antibody. Bottom panel: total amounts of the tested proteins (Input) analysed by western blotting with anti‐GFP or anti‐Myc antibody without immunoprecipitation. Lane 1, AtbZIP18; lane 2, AtbZIP52; lane 3, AtbZIP69; lane 4, AtposF21; lane 5, AtbZIP29; lane 6, AtbZIP30; lane 7, NtRSG; lane 8, AtVIP1; lane 9, free green fluorescent protein (GFP). Two independent experiments were performed for each assay with similar results.
