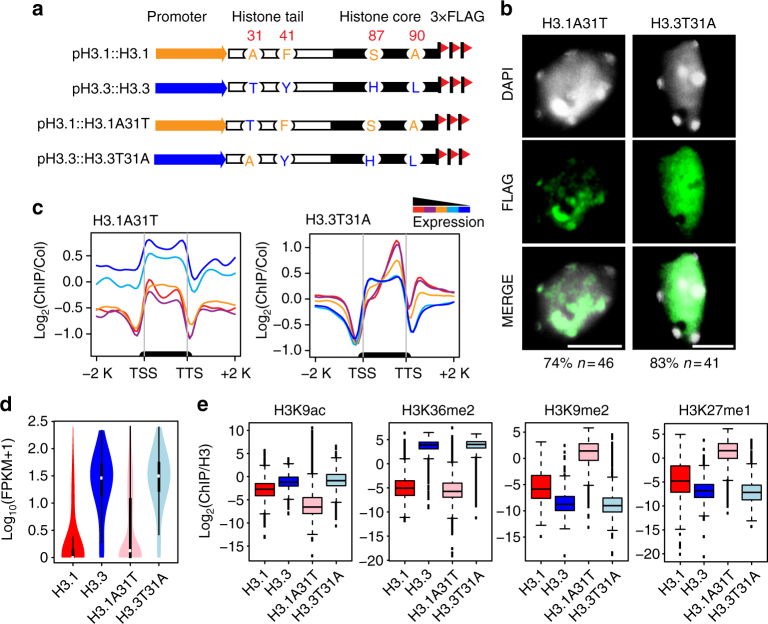
Fig. 4.
Ala/Thr31 is not decisive for genome-wide H3 distribution pattern. a Schematic diagram of H3.1, H3.3, H3.1A41T, and H3.3T31A FLAG-tagged constructs. b Localization of H3.1A31T and H3.3T31A protein in nuclei (bar = 5 μm). The n represents the total number of examined nuclei. The percentage describes the ratio of the nuclei showing the H3 distribution pattern out of total examined nuclei. DAPI indicates the DAPI staining of the nucleus. c Metagene plots of H3.1A31T and H3.3T31A ChIP-seq reads over five groups of genes divided based on their expression levels. The black bar in the x-axis represents genes. TSS transcription start sites, TTS transcription terminal sites; −2 K and +2 K represent 2 kb upstream of TSS and 2 kb downstream of TTS, respectively. The y-axis represents the log2 value of H3.1A31T and H3.3T31A ChIP-seq reads normalized to those of Col-0. d Violin plots of the expression levels of genes associated with H3.1, H3.3, H3.1A31T, and H3.3T31A ChIP-seq peaks. The y-axis represents the log10 value of FPKM +1. FPKM fragments per kilobase of transcript per million mapped reads. RNA-seq data were from 3-week-old seedlings. e Boxplots of histone modification levels in ChIP-seq peaks of H3.1, H3.3, H3.1A31T, and H3.3T31A. The y-axis represents the ChIP-seq reads normalized with those of H3. Histone modification ChIP-seq data were from 2-week-old aerial tissues