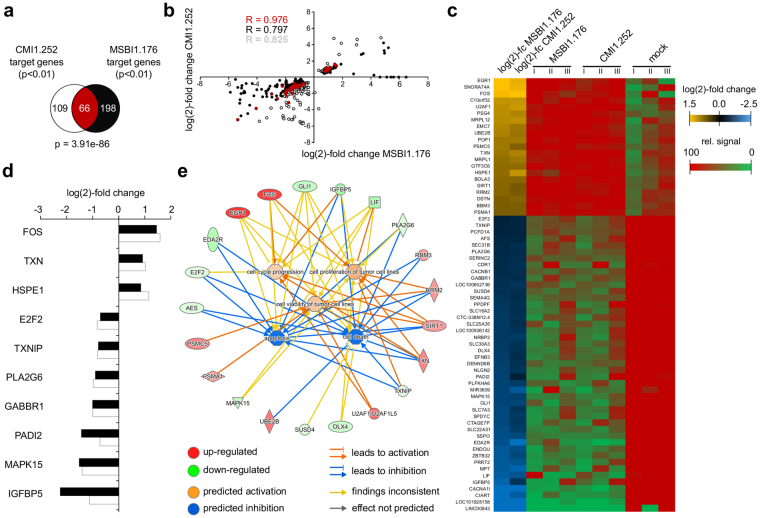
Figure 6.
Human gene expression changes upon MSBI1.176 and CMI1.252 expression in HEK293TT cells. (a)Venn diagram of the gene sets statistically significantly (p < 0.01) regulated upon MSBI1.176 expression (black) as well as upon CMI1.252 expression (white). The common subset is highlighted in red and the corresponding hypergeometric distribution p-value is given below. Gene numbers for each set are indicated. (b) Scatter plot of the genes shown in panel (a) using the same colors. The log(2)-fold change in expression upon MSBI1.176 expression is plotted on the x-axis and the log(2)-fold expression change upon CMI1.252 expression is plotted on the y-axis. The Pearson correlation coefficients (R) for the common subset as well as for the remaining genes of each single target gene set are indicated. (c) Heatmap of the common subset of MSBI1.176- and CMI1.252-regulated human genes. The log(2)-fold change in gene expression in both conditions (MSBI1.176 and CMI1.252 expression) is indicated as a color code for the common subset of 66 genes. Up-regulated genes are colored in yellow, down-regulated genes are colored in blue. For each gene, the relative, normalized signal for three biological replicates is indicated as a color code. (d) Representative genes co-regulated upon MSBI1.176 and CMI1.252 expression. Expression changes upon MSBI1.176 expression are colored in black, those upon CMI1.252 expression are colored in white. (e) Representation of genes involved in regulation of cell cycle progression, proliferation and viability of tumor cell lines, cell death and apoptosis as identified by canonical pathway enrichment analyses of the common subset shown in panels (a) and (c). Genes, whose expression is up-regulated upon BMMF expression are colored in red, those whose expression is down-regulated are colored in green.