Table 1.
Motifs and associated methylations present along the genomic DNA of S. acidocaldarius.
| Motif | Nber of motif in the genome | Nber of recognition sequences in the genome | Type of methylation | % of methylated motif |
|---|---|---|---|---|
| 5′-GGCC-3′ | 1,140 | 570
|
m4C | 97 |
| 5′-AGATCC-3′ | 1,022 | 1,022 | ||
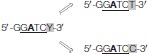 |
1,022 |  |
m6A | 24 |
| 578 | 289
|
m6A | 60.9 | |
| 5′-GATC-3′ | 2,622 | 1,311
|
m6A | 32.1 |
The SMRT sequencing revealed the presence of three motifs: 5′-GGCC-3′, 5′-AGATCC-3,′ and 5′-GGATCY-3′ with “Y” being a pyrimidine (“T” or “C”). The two latter motifs contain 5′-GATC-3′ as core motif (underlined in both) which can be studied as such. Modified nucleobases, for each motif, are written in bold. The number of motifs in the whole genome of S. acidocaldarius, the number of corresponding recognition sequences formed in the genome, the type of methylation and the percentage of methylated motifs are given in this table.
