Figure 6.
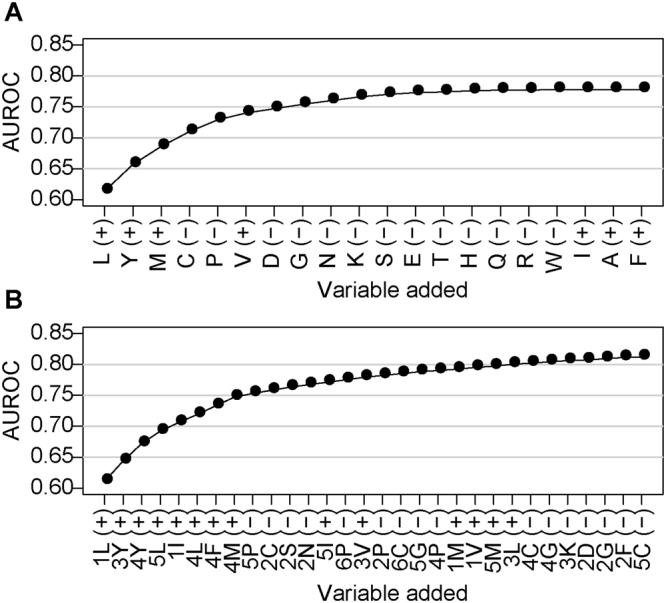
AUROC of the ADAMTS13 substrate motif. Logistic regression with forward stepwise feature selection was applied to ADAMTS13 selection of the VWF73(P3-P3′) library, with significantly cleaved peptides trained against neither significantly cleaved nor significantly depleted. This analysis provided a measure of which amino acid requirements were most useful for predicting ADAMTS13 substrate recognition, and how well a model based on the presence of these amino acids predicted peptide cleavage. Outcomes were iterated for amino acid regardless of position (A) or amino acids constrained to position (B). Both positive (+) and negative (−) regulators of ADAMTS13 substrate recognition were identified. Amino acid content alone yielded an Area Under Receiver Operating Curve (AUROC) of ~0.78 (A), whereas amino acid content at defined positions yielded a maximum AUROC of 0.8 (B).
