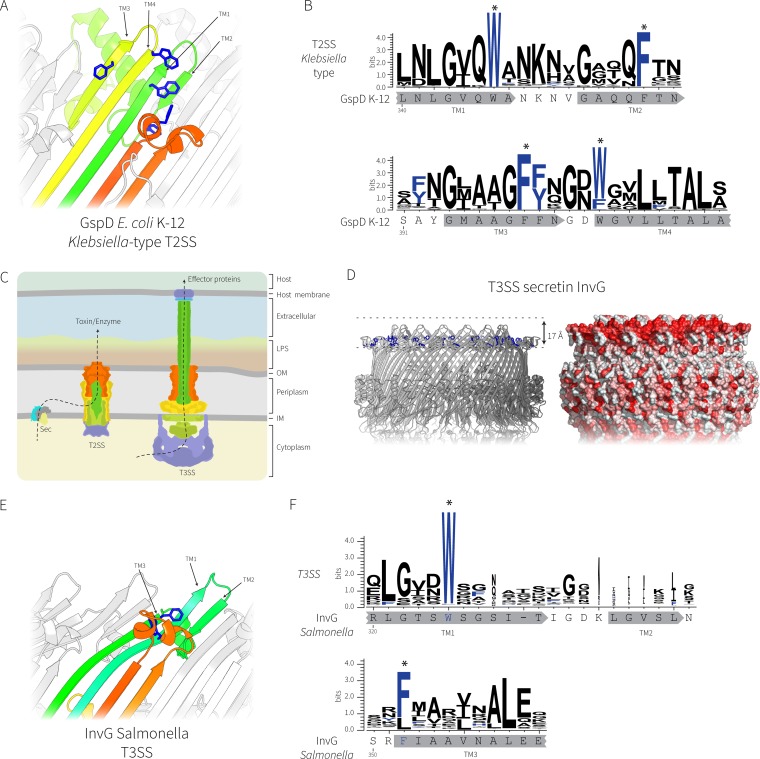
FIG 4.
Comparative analysis of the secretin complexes from T2SS and T3SS. (A) The transmembrane regions of the E. coli K-12 GspD secretin (PDB accession number 5WQ7) representing the Klebsiella-type T2SS secretin; the conserved aromatic residues are shown in blue. (B) Sequence logo showing the conservation of the aromatic residues (designated with asterisks) in the transmembrane β-strands was generated from 363 Klebsiella-type T2SS secretin sequences with the sequence from E. coli K-12 substrain DH5α shown for reference. The locations of the putative transmembrane β-strands are shown in gray below the sequence logos. (C) Cartoon depiction of the relationship between T2SS and T3SS, where the colors represent subunits with similar functions. LPS, lipopolysaccharide; OM, outer membrane; IM, inner membrane. (D) Similar transmembrane β-strands from the T3SS secretin InvG from Salmonella enterica (PDB accession number 5TCQ) with the hydrophobic belt shown. (E) The transmembrane region of InvG showing the conserved aromatic residues (blue). Note that the leftmost short β-strand is missing in the T3SS secretin, causing the upper lip of the barrel to curve inward to accommodate the missing strand. (F) Sequence logo of alignments of 1,543 T3SS secretins showing the conservation of the aromatic residues in the transmembrane β-strands. The InvG sequence and β-strands from Salmonella enterica are shown below for reference. The locations of the putative transmembrane β-strands are shown in gray below the sequence logos.